This article was originally published in the Breakthroughs Newsletter. Find more stories like this, as well as the new Breakthroughs Podcast, on the Breakthroughs homepage.
Imagine a marble, rolling down a hill. There are many paths the marble can take — many peaks and troughs guiding its descent to different places. As the marble moves down the hill, the paths and ultimate destinations available to the marble dwindle. The terrain drives where the marble lands.
This is how famed geneticist C.H. Waddington described his theory of “epigenetic landscape” for cell differentiation in 1957 — the marble is an uncommitted cell on the path of differentiation, the peaks and troughs are the pattern of gene expression that commits the cell to a certain identity. DNA methylation, a mechanism involved in a wide variety of epigenetic processes, wasn’t discovered until 1975, but Waddington’s initial premise held true.
Today, the study of epigenetics has taken off. In the last 20 years, laboratories across the world have been racing to identify molecular mechanisms of epigenetic regulation, finding they have an impact on more than just cell differentiation. Epigenetic mechanisms play a role in a huge range of processes, including oncogenesis and immune response, and more are discovered every year.
At Northwestern, the Simpson Querrey Center for Epigenetics (SQE) leads the way, connecting the Northwestern academic and medical community to integrate the study of epigenetics into science and clinical care.
It’s a living, breathing resource for scientists at Northwestern who want to learn more about changes in phenotype without a change in genotype, according to Ali Shilatifard, PhD, chair of Biochemistry and Molecular Genetics, the Robert Francis Furchgott Professor and director of the SQE.
“The main emphasis of this center is linking epigenetic discoveries — things that affect chromatin DNA and regulate gene expression — to human disease,” said Shilatifard, who is also a professor of Pediatrics. “Our colleagues throughout the medical center and university need expertise in this area, and now they have a place to go.”
A bi-weekly seminar called the Biochemistry Epigenetics and Metabolism Forum is a hub for the SQE, where members and non-members give updates on projects, share data and tout the use of core facilities co-sponsored by the center. There is also a monthly lecture series featuring experts in the field such as Robert Roeder, PhD (Rockefeller University), Mike Levine, PhD (Princeton University), Martine Roussel, PhD (St. Jude Children’s Research Hospital) and Lucy Godley, MD, PhD (University of Chicago).
In addition, the SQE will hold its inaugural symposium on November 30, bringing together scientists and clinicians from across the world for a day of presentations, discussion and networking. The SQE also plans to offer courses that will give investigators the tools they need to conduct epigenetics research, Shilatifard said.
“For example, we are planning to offer courses on different methods of RNA sequencing and data analyses, how to use ChIP-seq and related methodologies to identify functions of proteins that work on chromatin,” said Shilatifard, whose own work using these techniques has defined how chromosomal translocations cause pediatric cancers and how this information can be used to treat cancers. “We want to teach investigators how to fish, instead of simply giving them the fish.”
Pulmonary Patch-up
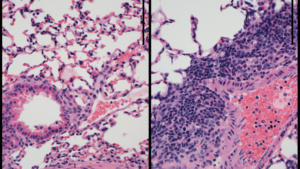
One area where epigenetics has been found to be influential is in immune response, specifically in lung repair.
Benjamin Singer, ’07 MD, ’11 GME, assistant professor of Medicine in the Division of Pulmonary and Critical Care and a member of the SQE, became interested in epigenetics’ effect on immune response during a fellowship at Johns Hopkins University School of Medicine.
“The complex cell types involved in lung repair are highly regulated by epigenetic mechanisms,” said Singer, who is also an assistant professor of Biochemistry and Molecular Genetics. “My main discovery was that by manipulating DNA methylation, we can actually encourage these cells to do their jobs better.”
Now with his own laboratory at Northwestern, Singer has continued to investigate the topic. In one study, published in the Journal of Biological Chemistry, Singer took a detailed look at DNA methylation in mouse models of neonatal lung infection.
With help from the SQE, Singer used bioinformatics and computational tools to show that DNA methylation patterns play an important role in controlling T-cells that help repair the lung. A subsequent paper, published in the Journal of Clinical Investigation Insight, used an innovative sampling technique to measure DNA methylation patterns in human patients with lung failure. Singer found that the patterns in humans were very similar to what they saw in models, opening the door for one day using these patterns to predict treatment response.
“If you could define a part of the genome that has this methylation, you might be able to predict the outcome,” Singer said. “One could draw blood from a patient, quickly assess the DNA methylation and estimate if this patient is likely to do poorly or to do well and respond to therapy.”
There is even potential for a drug-based therapy, if scientists can identify an actionable drug target.
“If you can manipulate DNA methylation, you could help patients get off the ventilator and make it more likely that they would survive this horrible illness,” Singer said.
These sampling and analyzing techniques, which will soon be expanded to a much larger group of patients as part of an ongoing study at Northwestern Medicine, are an example of how the SQE can help scientists at Northwestern.
“The SQE can affect research that spans from the atomic, molecular level all the way through application to patients,” Singer said. “This can’t be done at very many places — it makes this an incredible environment to do high-impact work.”
Onco-histones
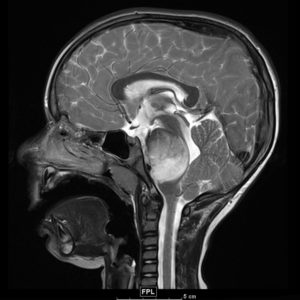
One of the most popular areas of epigenetics is oncogenesis. SQE member Amanda Saratsis, MD, ’14 GME, assistant professor of Neurological Surgery, became interested in diffuse intrinsic pontine glioma (DIPG), a rare and deadly pediatric tumor located in a part of the brainstem called the pons, during residency training at Georgetown University, when two groundbreaking papers discovered mutations in a protein called histone H3 in 80 percent of surveyed tumor samples.
“The presence of this histone mutation wreaks havoc on the epigenetic mechanisms of gene expression regulation in these tumors,” Saratsis said.
Since then, Saratsis has made it her mission to learn more about these tumors, working with Shilatifard and other collaborators at Northwestern to find their source and shut them down. In a paper published last year in Nature Medicine, Saratsis, Shilatifard and colleagues found that the mutation in histone H3 leads to one amino acid being swapped out for another, causing a downstream cascade that leads to tumor-promoting gene expression in the brainstem.
A child’s developing brain may be particularly susceptible to these epigenetic perturbations. For example, the genesis of these pontine tumors often coincides with a period of heavy myelination in the pons; the process by which neurons are insulated with a material called myelin. While myelin is needed to ensure information travels fast throughout the brain, the physiological signals that promote this normal process may also encourage DIPG cells to grow.
“We’re still not 100 percent sure what the cell of origin is for DIPG, but we think the rapidly developing brainstem may provide a unique microenvironment for tumor cells to respond to normal signals with abnormal biology,” Saratsis said.
In fact, epigenetic mechanisms may be more common in pediatric brain tumors than previously thought, Saratsis said.
“Now that we have this perspective, we’re going back with fresh eyes and looking at these epigenetic processes, and we’re finding abnormalities in pediatric brain tumors,” Saratsis said. “They don’t all have this histone H3 mutation, but they may have a different mutation that affects the same sort of process.”
Collaboration with other SQE members is crucial, especially for studying a disease like DIPG. Because these tumors are deep in the brainstem, high-quality specimens are hard to come by, so Saratsis is working with the SQE to develop new biological assays that can extract epigenetic information from archival tumor specimens or spinal fluid.
“The technology that the SQE provides allows us to approach these specimens that perhaps we would have no use for,” Saratsis said. “How we process specimens, perform sequencing and then analyze that data has proven very challenging in the past, and now we’re able to make gains like never before.”
The field of epigenetics is in its infancy, but the knowledge base and scientific techniques continue to grow and improve. The top-to-bottom integration into both Singer’s and Saratsis’ research is something the SQE hopes to foster at Northwestern, galvanizing scientists across institutions and centers to produce solutions for patients.
“The beauty of Northwestern and the Simpson Querrey Center for Epigenetics is that we can go from basic chemistry, to biochemistry, to molecular genetics, into animal models and finally into the clinic, all at one institution,” Shilatifard said.
Find more information about SQE education events and seminars here.
Shilatifard and Saratsis are also both members of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University, and Shilatifard is co-leader of its Epigenetics and Nuclear Dynamics Program.