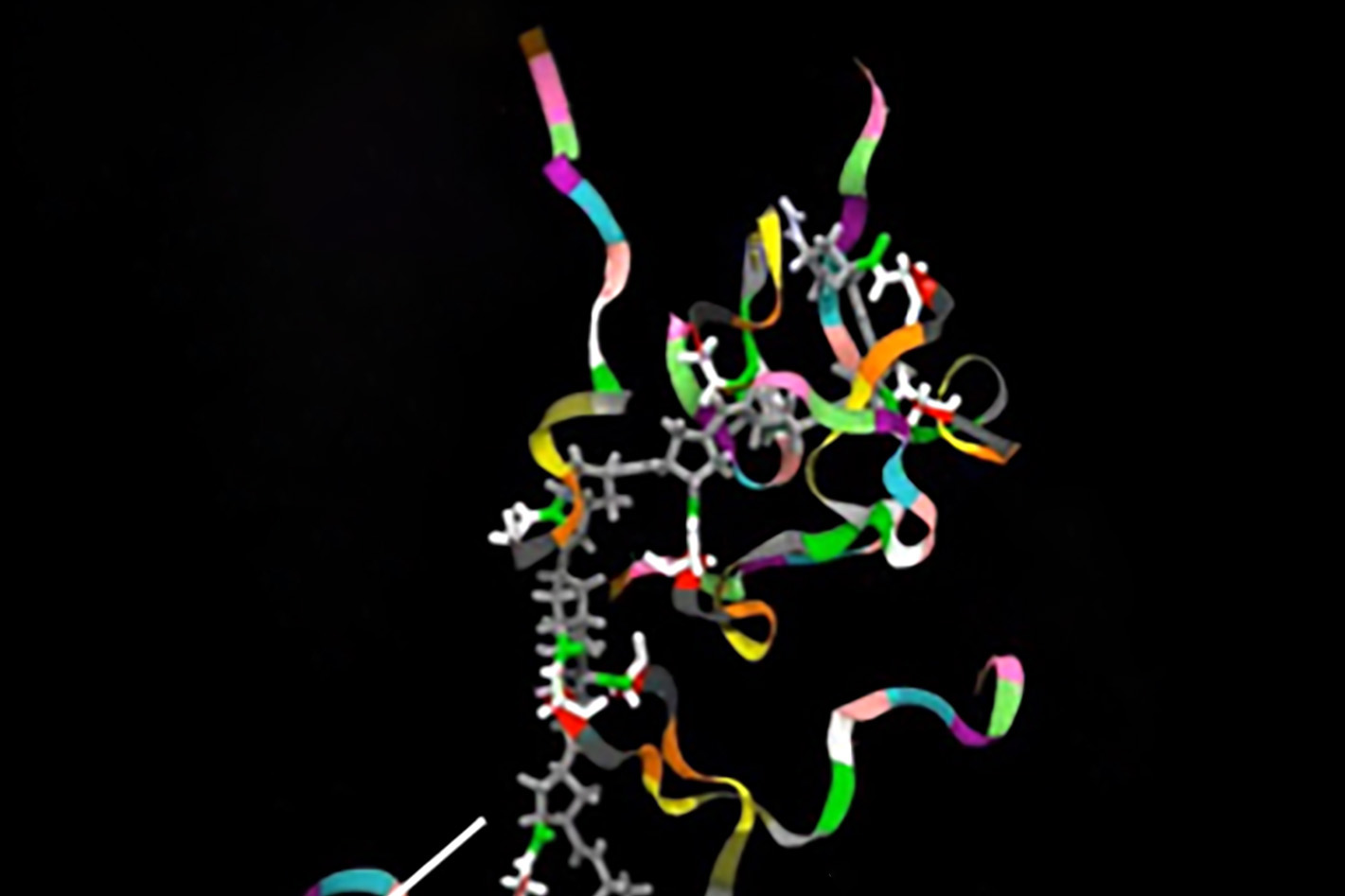
Northwestern Medicine scientists have identified a new gene that can inhibit a multi-protein complex, possibly increasing the risk of cancer, according to a new study published in Science Advances.
The gene, called CATACOMB, is activated by epigenetic mechanisms and binds to a subunit of polycomb repressive complex 2 (PRC2), a protein complex known for its involvement in cancer. When CATACOMB is bound to PRC2, the entire complex stops working and this mechanism can cause cancer, according to Ali Shilatifard, PhD, chair of Biochemistry and Molecular Genetics, director of the Simpson Querrey Center for Epigenetics and senior author of the study.
“PRC2 is a protein complex mutated in a large number of cancers – it’s a major player,” said Shilatifard, who is also Robert Francis Furchgott Professor, a professor of Pediatrics and leader of the Cancer Epigenetics & Nuclear Dynamics (CEND) program at the Robert H. Lurie Comprehensive Cancer Center of Northwestern University.

Polycomb-group protein complexes play a fundamental role in several physiological processes due to their ability to suppress gene transcription. Transcription, the first step of gene expression, is the process of reading instructions from a genome and producing messages used to synthesize proteins or other molecules necessary for life. In particular, some mutations that are known to activate PRC2 have been identified as drivers of cancer: when PRC2 is misregulated, many important target genes can be aberrantly silenced and cancer cells can use this to replicate more quickly.
Andrea Piunti, PhD, postdoctoral fellow in the Shilatifard laboratory and lead author of the study, was investigating interactions between PRC2 and other molecules when he discovered CATACOMB and its connection to PRC2. Curiously, CATACOMB was dormant in every cell type Piunti examined except for a few types, including osteosarcoma cancer cells.
Searching for some common feature of CATACOMB-dormant cells, he found they all had high levels of DNA methylation, an epigenetic mechanism that cells use to shut off certain genes from being expressed. On the other hand, other cells used in the experiments had low levels of DNA methylation, and CATACOMB was expressed, binding to PRC2 and disabling it.
To find out if DNA methylation was the key, Piunti deployed a DNA methylate inhibitor on the CATACOMB-dormant cells, finding that once methylation was lowered, CATACOMB was expressed similarly to the osteosarcoma cells.

According to Piunti, this means the regular function of CATACOMB may be as a sensor of DNA methylation: varying levels of DNA methylation are required at different periods of development or during disease, and activation of CATACOMB may play an important role in these settings.
In addition, CATACOMB bears similarities to a 3K27M histone mutation frequently present in pediatric high-grade brain tumors, according to the authors.
In the future, Shilatifard and Piunti plan to explore therapeutic applications of CATACOMB. In addition, the fact that CATACOMB is silenced in regular cells but present in germ cells and placenta is another peculiarity worthy of further investigation, Shilatifard said.
“A lot about CATACOMB has been answered by this paper, and even more questions have been raised,” Shilatifard said.
This work was supported by grants from the National Institutes of Health; R00DC013805, K08HL128867, R50CA211428, R37-HD38691 and R35CA197569.