Northwestern Medicine investigators have identified a previously unknown regulator of tumor immune evasion, which may help improve the efficacy of current and future anti-tumor immunotherapies, according to recent findings published in the Journal of Clinical Investigation.
“The study provides a molecular insight into understanding why some cancer patients cannot be treated by the checkpoint blockade antitumor therapy, but others can,” said Deyu Fang, PhD, the Hosmer Allen Johnson Professor of Pathology and senior author of the study.
Antitumor immunotherapy is a type of cancer treatment that helps the immune system in fighting cancer and includes a range of therapy types, such as immune checkpoint inhibitors. Immune checkpoints help prevent the immune system from being too strong and eradicating other cells, including cancer cells.
By targeting these checkpoints using checkpoint inhibitor drugs, the immune system can better respond to and fight off cancer cells. However, not all patients will respond well to immunotherapy and the underlying reason has remained unclear.
“The big question is whether we can find a better approach to make this therapy work for all patients,” Fang said.
A common immune checkpoint protein targeted by anti-tumor immunotherapy drugs is PD-L1, which is expressed on the surface of immune cells and is also increased on the surface of certain cancer cells, helping them evade the immune system.
Therefore, identifying novel regulators of PD-L1 expression in tumors may improve the efficacy of antitumor immunotherapies, according to Fang.
In the current study, Fang’s team developed a CRISPR-based screening platform to analyze the entire family of deubiquitination genes from both mice and human PD-L1 lung cancer cell lines. Using this approach, the investigators discovered that the ATXN3 gene promotes tumor immune evasion by promoting PD-L1 expression in tumor cells at the transcriptional level.
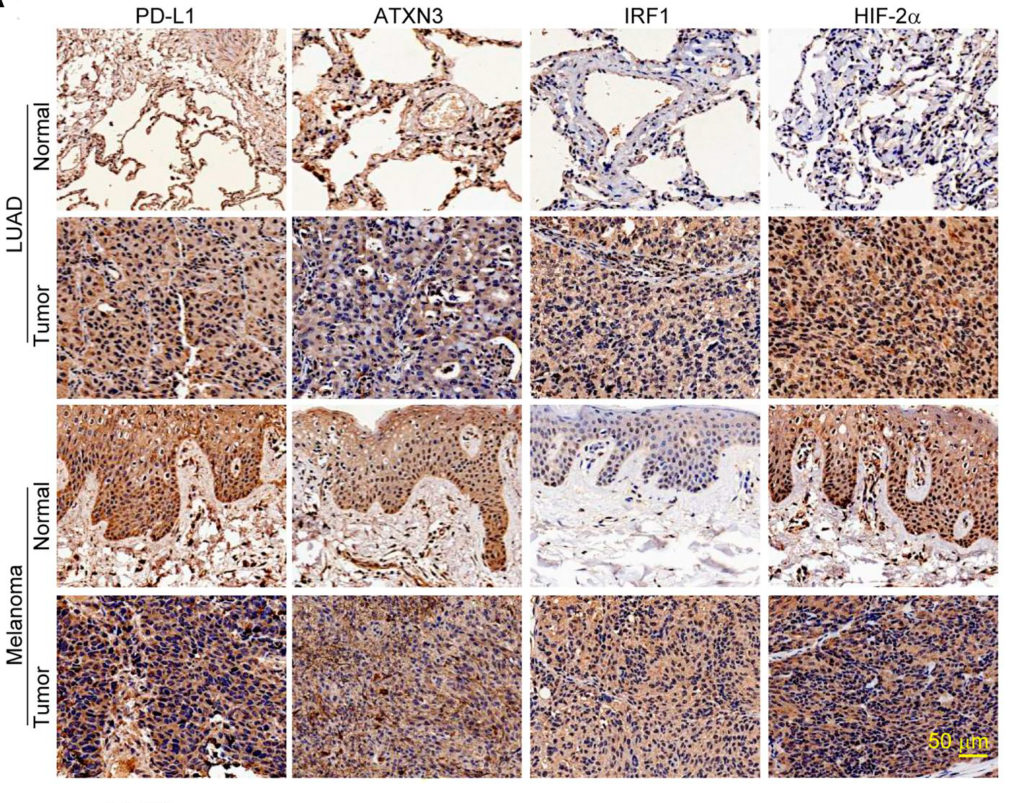
Further analysis using The Cancer Genome Atlas database revealed a positive correlation between the genes ATXN3 and CD274 — which encodes PD-L1 — in more than 80 percent of human cancers. Notably, ATXN3 was positively correlated with PD-L1 expression and its transcription factors in lung adenocarcinoma, the most common type of non-small cell lung cancer, and melanoma.
“Since ATNX3 promotes PD-L1 expression, we posed that ATXN3 suppression may enhance antitumor immunity in vivo,” said Fang, who is also a member of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University.
Using CRISPR and other targeted gene expression techniques to knockout ATXN3 in mouse models of PD-L1 lung carcinoma, the investigators found that suppressed ATXN3 enhanced antitumor immunity in the mice and improved the efficacy of PD-1 antibody therapy.
The findings suggest that ATXN3 is a positive regulator for PD-L1 tumor expression and tumor immune evasion. According to Fang, the findings also suggest that selectively targeting ATXN3 may improve the efficacy of antitumor immunotherapies as well as reduce toxicity and adverse side effects for all patients.
“If we combine an ATXN3 inhibitor and the current anti-tumor immunotherapy, we can improve the therapeutic efficacy and reduce the amount of antibody needed, meaning reduce the side effects,” Fang said.
This work was supported by National Institutes of Health grants R01DK126908, R01DK120330, R01CA257520 and CA232347, the Shandong Province Natural Science Foundation, the National Natural Science Foundation of China and the Dalian High-Level Talent Innovation Support Program.