The DNA methylation landscape of regulatory T-cells is more complex than previously understood, according to a new study published in The Journal of Clinical Investigation.
These findings could inform the development of regulatory T-cell (T-reg) therapies for cancer and autoimmune diseases, according to Benjamin Singer, ’07 MD, ’10 GME, assistant professor of Medicine in the Division of Pulmonary and Critical Care and senior author of the study.
“This is another piece of the puzzle as people think about designing therapeutics to leverage epigenetic mechanisms like DNA methylation,” said Singer, who is also an assistant professor of Biochemistry and Molecular Genetics.

T-reg cells are counter-regulatory cells that tamp down immune system activation to prevent damaging healthy cells. T-reg cells that function incorrectly can be a contributor to auto-immune diseases such as multiple sclerosis and rheumatoid arthritis.
“They are responsible for keeping the immune system in check so things don’t go haywire,” Singer said.
On the other hand, normally functioning T-reg cells can complicate matters in some situations. In cancer, T-reg cells can facilitate cancer growth by reducing immune activity. Immunotherapy, an emerging cancer treatment, takes advantage of this function by blocking T-reg cells’ ability to reduce inflammation, unleashing the full power of the immune system against the cancer.
One class of drugs, DNA methyltransferase inhibitors, affects T-reg cells by modifying DNA methylation patterns associated with this inflammation reduction. DNA methylation acts as a muffler on the genome: sites that are methylated are less likely to be expressed.
Until now, the only role for methylation in T-reg cells that was firmly established was a lack of methylation — called hypomethylation — at specific sites on the genome, resulting in expression at those sites.
However, Singer and his colleagues were interested in other sites that did have methylation, he said.
“Do these other sites need that maintenance DNA methylation to keep a T-reg cell a T-reg?” Singer said, describing the research question.
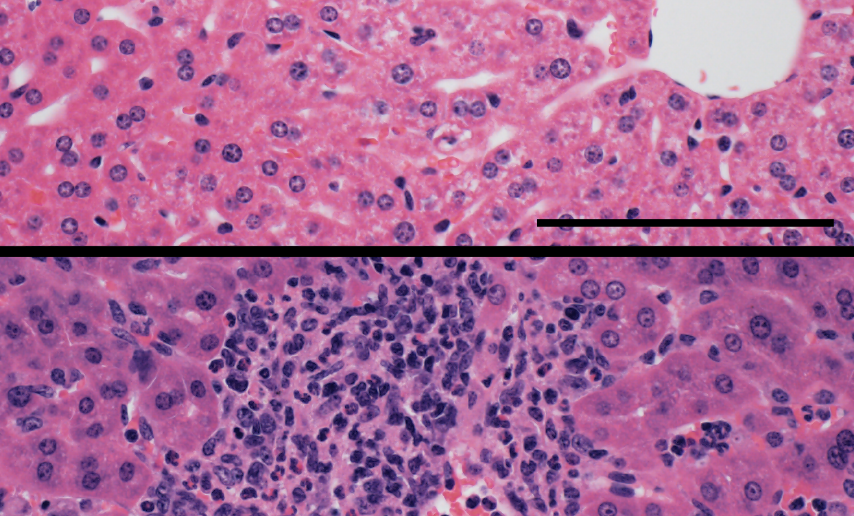
In the study, investigators disabled a key molecule involved in DNA methylation in mouse models, both from birth and in a manner that allowed scientists to deactivate the molecule once the mouse was fully grown. In both cases, inflammation ran rampant in the mice after the molecule was turned off.
Genetic analysis revealed that many of the normally methylated sites coded for inflammatory programs; functions belonging to other immune cells, but not T-regs.
“T-reg cells silence or repress those parts of the genome because that’s not part of their T-reg identity,” Singer said. “If those sites are not methylated, those T-regs can become hyper-inflammatory and cause pathology.”
This is relevant for DNA methyltransferase inhibitors, some of which reduce methylation across the board in T-reg cells. Singer said he believes a more targeted approach may be required in order to bring these therapies to their full potential.
“In some cases, knocking down methylation could have beneficial effects,” Singer said. “But you really do need to maintain methylation at certain places in the T-reg cell genome, otherwise you can unleash inflammation and cause problems.”
This study was supported by National Institutes of Health grants T32HL076139, F32HL151127, K08HL128867, U19AI135964, R01HL149883 and T32GM008152. The study was also supported by the David W. Cugell and Christina Enroth-Cugell Fellowship and the Francis Family Foundation’s Parker B. Francis Research Opportunity Award.