Using a novel integrative computational technique, scientists from Northwestern and the University of Illinois at Urbana-Champaign (UIUC) were able to classify disease conditions at the molecular level using epigenomic data sets, according to a recent study published in Cell Reports.
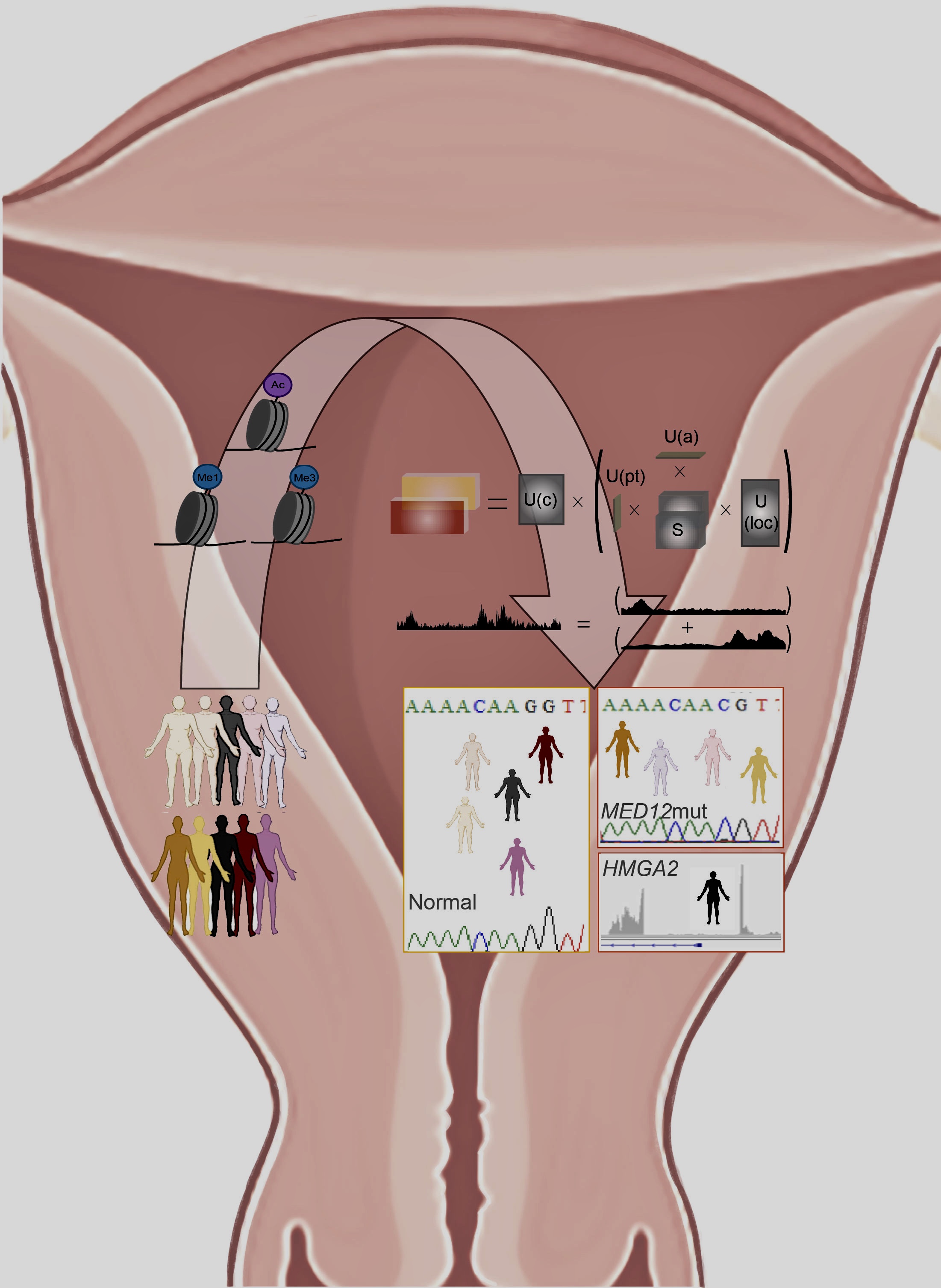
The new approach, collaboratively developed by the authors from Northwestern and UIUC, is called DeCET (Decomposition and Classification of Epigenomic Tensors); it analyzes complex, heterogeneous data to identify epigenomic differences between tissue types, disease subtypes, and changes in cell type, or cellular differentiation, explained Debabrata Chakravarti, PhD, vice chair for translational research and the Anna Lapham Professor of Obstetrics and Gynecology and lead author of the study.
The epigenome — the collection of all modifications to histone proteins and genome that alter gene expression — carries essential instructions for specifying cellular identity, so therefore epigenomic assays can offer a robust diagnostic marker for a wide range of diseases, according to Chakravarti, also a professor of Pharmacology and assistant director of shared resources at the Robert H. Lurie Comprehensive Cancer Center. For example, the pattern of DNA methylation can classify primary and secondary central nervous system tumors, as well as identify the cell type of a cancer’s origin.
Jun Song, PhD, Founder Professor of Physics at UIUC and Grant Barish, MD, associate professor of Medicine in the Division of Endocrinology at Northwestern, were co-senior authors of the study, and Priyanka Saini, PhD, of Northwestern and Jacob Leistico of UIUC, were co-first authors of the study. Leistico led the algorithmic development, and Saini led the experimental work, building a synergistic team between the two institutions.

The collaborative team of basic scientists, clinicians, and physicists conducted initial studies on uterine fibroids, or leiomyomas, which are non-cancerous tumors of uterine smooth muscle cells with limited treatment options that develop in up to 70 percent of women.
Scientists first performed epigenomic analysis of 25 normal uterine and 25 matched fibroid samples from patients. The team then used DeCET to analyze the data and identify key epigenomic features that could clearly distinguish normal myometrium tissue from leiomyoma disease states and disease subtypes.
Investigators next applied DeCET to “unknown” human samples to predict tissue status. The methodology accurately discriminated between normal and disease types and subtypes using the epigenomic signatures.
Finally, to demonstrate the general applicability of DeCET, the team extended their analysis using publicly available cancer epigenomic data sets and successfully classified other diseases, such as breast and prostate cancer.
“Our studies overcome drawbacks of current analysis methods, such as arbitrary choices of meta-analysis parameters and inter-sample variability,” explained Saini and Leistico.
DeCET also outperformed existing tools in stratifying distinct tissue types, disease subtypes and cellular differentiation states, according to Barish.
“Epigenomic profiling is a path towards personalized medicine,” Chakravarti said.
“We expect that our study will help better understand and identify disease states, thereby facilitating future therapeutic, diagnostic and prognostic strategies,” added Song, who supervised the computational algorithmic development.
Other Northwestern co-authors included: Serdar E. Bulun, MD, the Chair and John J. Sciarra Professor of Obstetrics and Gynecology; Jian-Jun Wei, MD, the Floyd Elroy Patterson Research Professor of Pathology in the Division of Gynecologic Pathology and director of gynecologic pathology in the Department of Pathology; J. Brandon Parker, PhD, research assistant professor of Obstetrics and Gynecology in the Division of Reproductive Science in Medicine; Magdy Milad, MD, MS, the Chief and Albert B. Gerbie, MD, Professor of Obstetrics and Gynecology in the Division of Minimally Invasive Gynecologic Surgery; Poorva Sandlesh, PhD, postdoctoral fellow in the Chakravarti Laboratory; Christopher Futtner, PhD, research associate, and Yasuhiro Omura, research technician, in the Barish Laboratory; and Pritin Soni, research technician in the Chakravarti Laboratory.
Miroslav Hejna, PhD, postdoctoral fellow in the Song Laboratory at UIUC, worked with Leistico to develop and test the tensor classification algorithm.
This study was supported by National Institutes of Health grants R01HD089552, P01HD057877, P50HD098580, R01CA163336 and R01CA196270.