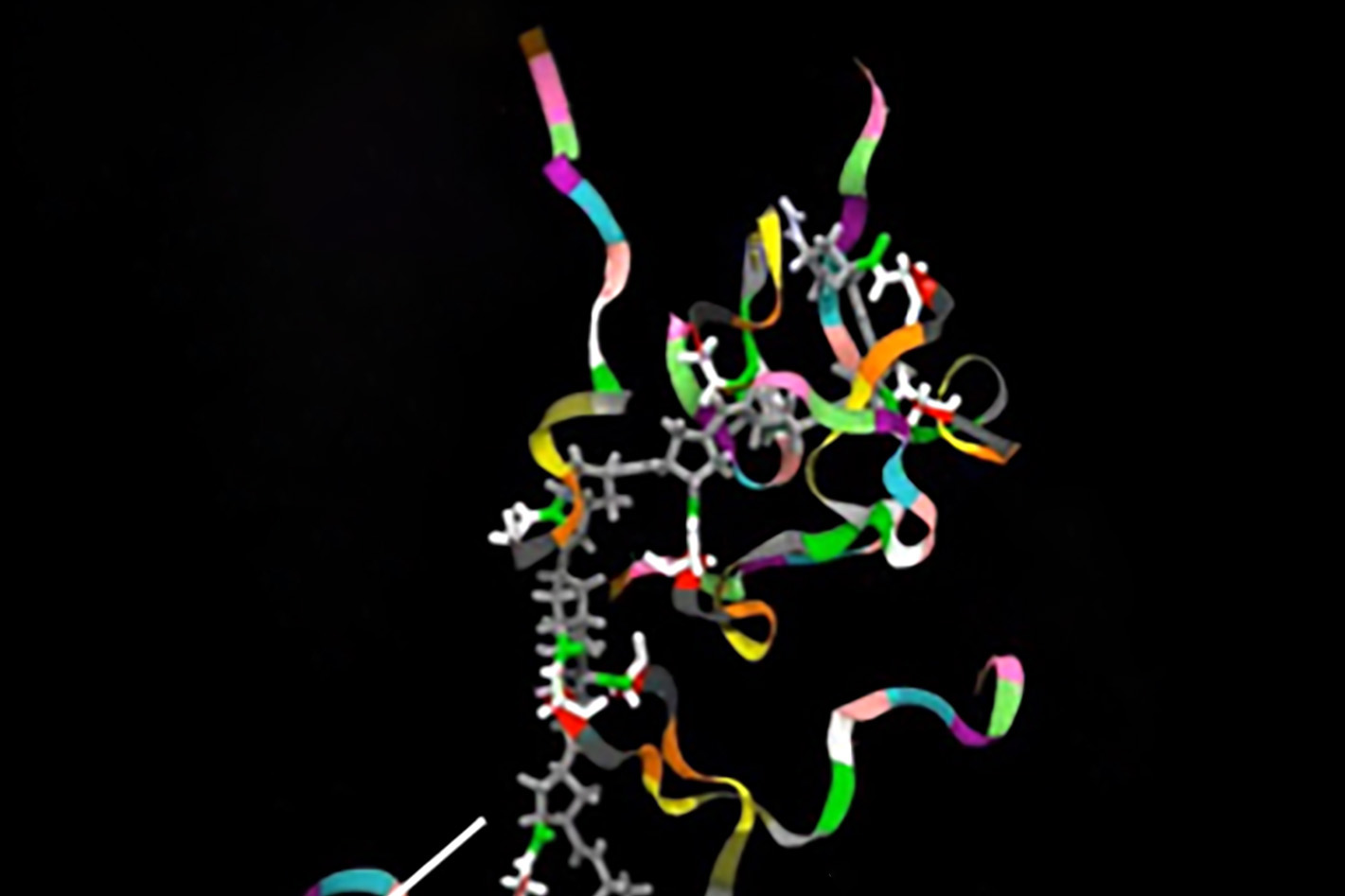
Northwestern Medicine scientists have detailed the diverse function of CDK9-containing complexes and their impact on gene expression in a recent study published in the journal Genes and Development. The findings shed light on potential pathway inhibitors that could be the target of novel cancer therapies, according to Ali Shilatifard, PhD, the Robert Francis Furchgott Professor, chair of Biochemistry and Molecular Genetics and senior author of the study.
“If you inhibit CDK9, you’re going to inhibit all of these pathways” said Shilatifard who is also a professor of Pediatrics, director of the Simpson Querrey Institute for Epigenetics and a member of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University.
RNA polymerase II catalyzes gene expression, which is regulated by various transcription factors to ensure proper cell growth, differentiation and survival during periods of stress. The protein kinase CDK9, in particular, is a center for transcriptional regulation and plays a key role in this process.
“RNA polymerase II is regulated at the step of its release from promoter-proximal pausing and this release depends on the catalytic activity of CDK9, with its inhibition blocking the transcription of thousands of genes,” said Bin Zheng, a fourth-year student in the Driskill Graduate Program in Life Sciences and first author of the study.
Previous studies have uncovered multiple CDK9 protein complexes in humans, including BRD4-CDK9 and the CDK9-containing super elongation complex, or SEC. For the current study, the scientists aimed to understand the roles of these two complexes when under normal conditions and in response to cellular stress.
To determine the function of each complex, the investigators combined gene editing technology and protein inhibitors; an acute-depletion strategy called auxin-inducible degron (AID) and small molecule inhibitors of bromodomain and extraterminal domain (BET) proteins and SEC family proteins. AID allows for the use of CRISPR gene editing to tag and deplete any select protein, while the inhibitor and degrader simultaneously targeted related proteins and homologs.

“We had the potential to determine redundant and non-redundant roles of these different family members, a challenge when studying complex organisms such as humans,” Zheng said.
According to Zheng, the study highlighted the varying roles these factors play in regulating gene expression and may pave the way for the future development of more targeted therapies, including those that may help provide lower toxicity when used to treat associated cancers, like MLL-rearranged leukemia.
As for next steps, the investigators hope to uncover whether BRD4 and SEC are responsible for the hypoxia, when cell oxygen levels decrease due to stress, and other rapid transcriptional responses —which all may also occur during rapid tumor cell growth.
This work was supported by the Japan Society for the Promotion of Science Research Fellowship for Young Scientists, the Uehara Memorial Foundation Research Fellowship and the National Institutes of Health grants R50CA211428 and R01CA214035.