A multi-institutional team of investigators has developed a new framework for supporting healthcare providers in implementing polygenic risk score-based testing in primary care settings, according to a recent study published in Nature Medicine.
“This is an important step towards using genetic information to provide better healthcare for all,” said Elizabeth McNally, MD, PhD, the Elizabeth J. Ward Professor of Genetic Medicine and director of the Center for Genetic Medicine, who is also a co-principal investigator of Northwestern’s eMERGE clinical site and a co-author of the study.

“We’ve known that many common diseases have genetic influence, and now we can define genetic risk and use that information to tailor health recommendations and management. A key part of the study was ensuring that the genetic assessment worked in many different groups of people, and we were able to do that,” McNally said.
Polygenic risk scores (PRSs) are numbers, or scores, that measure how likely someone will develop a specific disease based on their genetic makeup. However, implementing PRSs into clinical care and interpreting and communicating results to both providers and diverse patient populations has been an ongoing challenge.
“PRSs are also known to have a lot of population specificity, so it could introduce a lot of disparity because a polygenic risk score developed in essentially a population that’s exclusively white won’t perform as well in people of other ancestries or backgrounds,” said Adam Gordon, PhD, assistant professor of Pharmacology, who led the prostate cancer arm of the study.
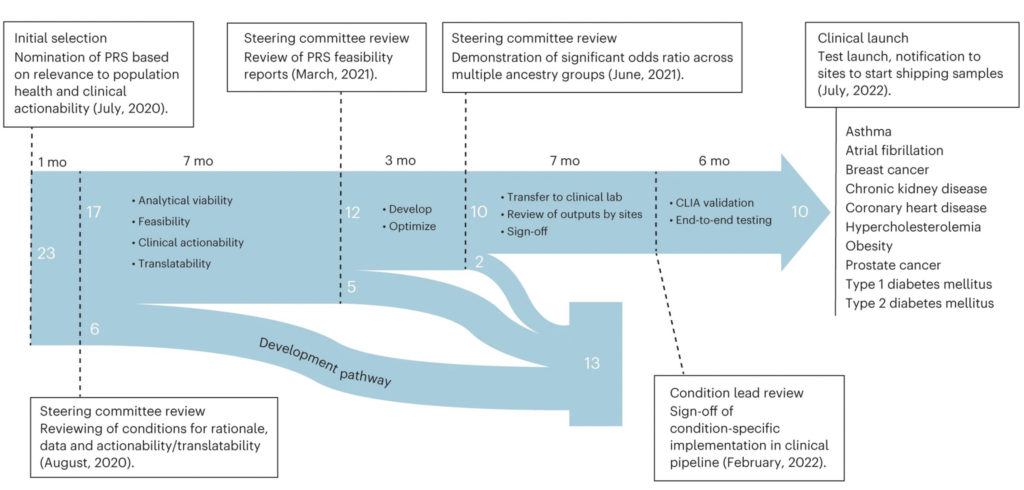
In response, the National Human Genome Research Institute-funded Electronic Medical Records and Genomics (eMERGE) Network — with Northwestern being one of more than 10 eMERGE clinical sites across the United States — developed a framework to evaluate and optimize PRS-based risk assessments of 25,000 adult and children eMERGE participants.
In the current study, the investigators selected preexisting PRSs from the Polygenic Score Catalog database and validated the scores with existing eMERGE data.

“PRS sum together genetic data from an individual and not all PRSs are created equal. There can be multiple PRSs for a condition that include different genetic markers. For this study, we needed to identify the best-performing PRSs while also paying attention to ease of implementation,” said Megan Roy-Puckelwartz, PhD, assistant professor of Pharmacology who led the atrial fibrillation (AFib) arm of the trial.
The investigators then chose 10 chronic diseases from an initial list of 23 conditions for implementation based on previous PRS performance, medical actionability and potential clinical utility, including cardiometabolic diseases and cancer. The selected chronic diseases included: asthma, AFib, breast cancer, chronic kidney disease, coronary heart disease, hypercholesterolemia, obesity, prostate cancer, type 1 diabetes and type 2 diabetes.
The investigators then developed a pipeline for clinical PRS implementation, which involved recommendations for transferring scores to a clinical laboratory and parameters for the validation and verification of score performance. They also used genetically diverse data from 13,475 participants enrolled in the All of Us Research Program to train and test their newly developed model.

Lastly, the team created a framework for regulatory compliance and developed a PRS clinical report for healthcare providers and to include in an additional genome-informed risk assessment.
The goal, Gordon said, is to move PRSs into the primary care setting in a way that providers find them efficient and easy to use among diverse patient populations.
“Primary care physicians already have a lot on their plate and adding a new kind of genomic score that might be difficult to interpret may not always be the best decision. However, if we can make it easy enough to use, it could be a really useful tool in identifying people who can’t be identified through other risk factors,” Gordon said.
Co-authors of the study include Lorenzo Pesce, PhD, research assistant professor of Pharmacology, Megan Roy-Puckelwartz, PhD, assistant professor of Pharmacology, Theresa Walunas, PhD, associate professor of Medicine in the Division of General Internal Medicine, and Yuan Luo, PhD, associate professor of Preventive Medicine and of Pediatrics, and chief AI officer for the Northwestern Clinical and Translational Sciences (NUCATS) Institute and the Institute for Artificial Intelligence in Medicine (I.AIM).
This work was partially supported by the National Human Genome Research Institute through grant U01HG011169.