In a study published in eLife, Northwestern scientists determined how two protein mutations which cause impaired motor function in Parkinson’s disease independently disrupt neuron activity.

The research is the first comparative approach examining multiple mutations in the protein LRRK2 side-by-side and was a collaboration between Loukia Parisiadou, PhD, assistant professor of Pharmacology, and Yevgenia Kozorovitskiy, PhD, the Soretta and Henry Shapiro Research Professor of Molecular Biology and associate professor of Neurobiology at the Weinberg College of Arts and Sciences.
Parkinson’s disease symptoms — including tremor, movement and stiffness — result from the degeneration of dopamine-producing neurons located in the midbrain. Striatal spiny projection neurons (SPN) that sense dopamine strongly express LRRK2. Together with the dopamine-producing cells, they form a brain network critical for motor control and reward systems.
Although Parkinson’s disease is classically considered a spontaneously emerging disease, it is now recognized to have a substantial genetic component, explained Parisiadou. The team focused on the two most common pathogenic mutations of LRRK2 — R1441C and G2019S.
“Our findings suggest that mutations in LRRK2 disrupt neuronal activity in the striatum before the loss of dopamine signaling,” Parisiadou said.
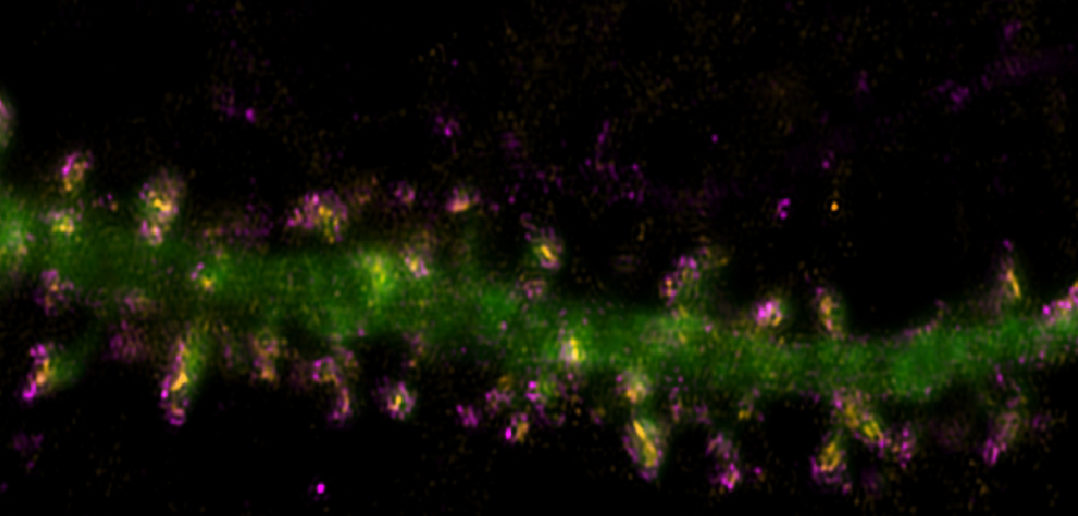
Scientists observed that the LRRK2 mutations reshaped the structure and function of SPN synapses, disrupting signals to other brain regions both via direct pathways, which promote movement, and indirect pathways, which suppress movement.
The confirmation of pre-clinical, pre-symptomatic changes caused by LRRK2 mutations suggests a potential therapeutic target.
“This work opens the possibility of developing individualized therapies aimed at supporting SPN health in Parkinson’s disease-related genetic mutation carriers before symptoms of dopamine loss arise,” Parisiadou said.
Using mouse models, the team utilized molecular, anatomical and electrophysiological approaches to examine LRRK2-driven dysfunctions on both a global, systematic scale and at the level of single synapse-specific effects.
The scientists determined that the LRRK2 R1441C mutation plays a critical role at the individual synapse level, altering the structure and function of SPN connections, particularly in the direct pathway.
Additional Northwestern study authors include Parisiadou lab members Chuyu Chen, PhD, and Shuo Kang, PhD; and Kozorovitskiy lab members Giulia Soto, Vasin Dumrongprechachan, and Nicholas Bannon, PhD.
Loukia Parisiadou was supported by National Institutes of Health (NIH) National Institute of Neurological Disorders and Stroke (NINDS) grant R01NS097901 and the Michael J. Fox Foundation for Parkinson’s Research LRRK2 Challenge. Yevgenia Kozorovitskiy was funded by NINDS R01NS107539, the Rita Allen Foundation Scholar Award, Kinship Foundation Searle Scholar Award (YK) and Arnold and Mabel Beckman Young Investigator Award. Nicholas Bannon, PhD, was supported by NINDS F32 NS103243. Vasin Dumrongprechachan was supported by a predoctoral award from the American Heart Association (19PRE34380056). SIM imaging was performed at the Center for Advanced Microcopy, supported by NIH 1S10OD016342-01 and NCI CCSG P30 CA060553.