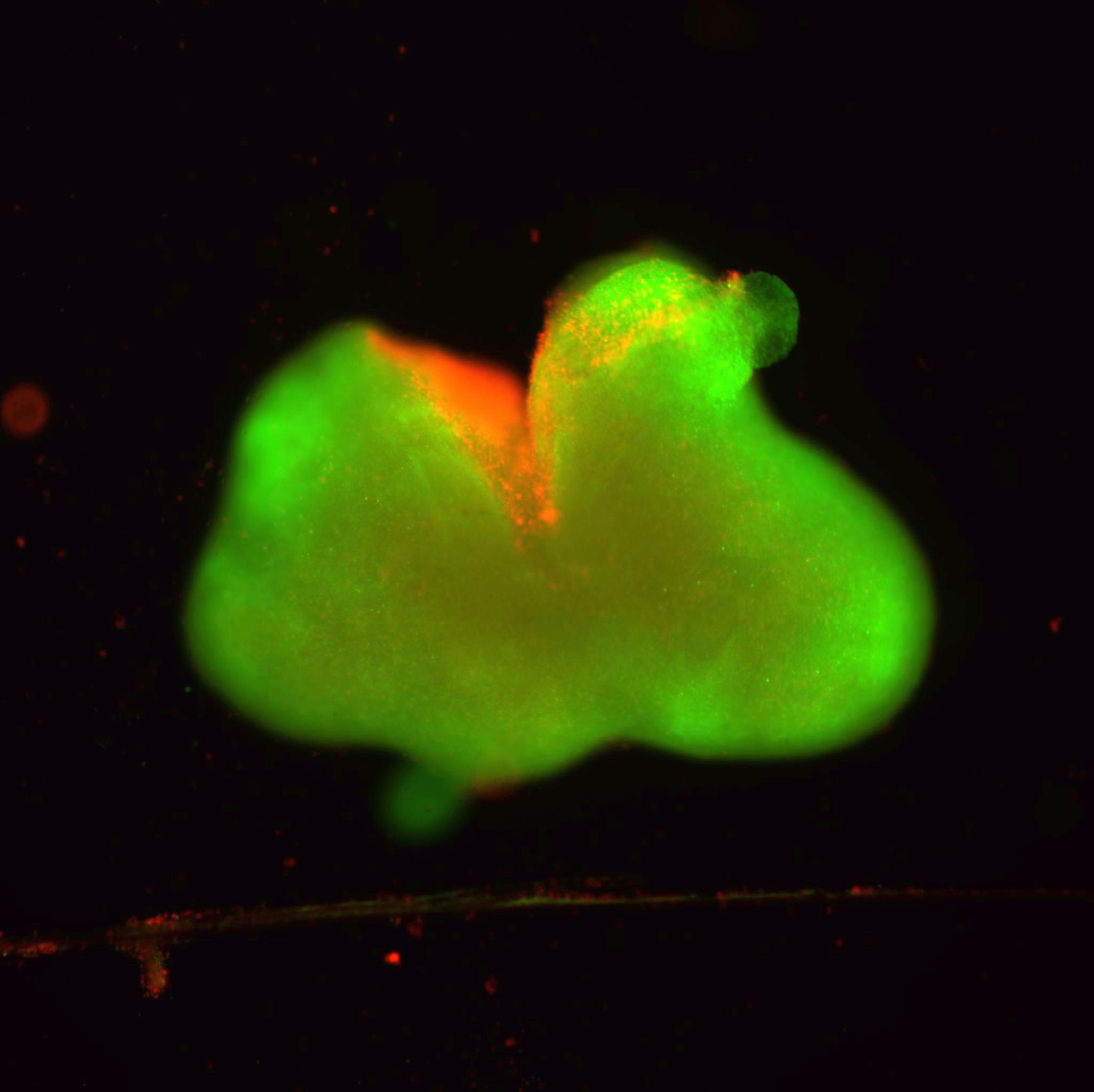
Cancer doesn’t just hijack the body’s cells — in the case of T-cell acute lymphoblastic leukemia, it physically alters the architecture of the genome, bringing together different genetic “neighborhoods” to fuel the replication and spread of cancerous cells, according to a Northwestern Medicine study published in Nature Genetics.
There is a silver lining, however: these changes might be targetable with small molecule drugs, a strategy that could fight cancer while sparing healthy cells, according to Panagiotis Ntziachristos, PhD, assistant professor of Biochemistry and Molecular Genetics and co-lead author of the study.
“These oncogenic neighborhoods might be more sensitive to small molecule inhibitors compared to the DNA in physiologically normal cells,” said Ntziachristos, also an assistant professor of Medicine in the Division of Hematology and Oncology and a member of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University. “The impact of these findings in the clinic could be substantial.”
DNA is organized into domains or “neighborhoods,” areas of chromatin with high genetic connectivity. In normal cells, these neighborhoods are bookended by insulator proteins. In certain cancers, such as T-cell acute lymphoblastic leukemia, those insulator protein binding sites are altered.
In the current study, Ntziachristos and his collaborators used patient-derived cells and advanced imaging and molecular biology technologies to investigate how insulator protein binding is altered in this form of leukemia. They found that the common cancer-causing gene MYC presents with a new neighborhood formation due to the activity of NOTCH1 transcription factor and altered binding of insulator proteins, allowing for aberrant MYC expression in cancer.
“This allows for genes and genetic control elements, such as gene enhancers, to be in close proximity, compared to normal settings where they are not close together,” Ntziachristos said. “This leads to aberrant activation of genes, an event that might contribute to oncogenesis.”
The investigators also applied a NOTCH1 inhibitor to patient-derived cancer cells, but found the inhibitor did not adequately reverse the formation of the MYC neighborhood to impact tumor growth.
“This might explain why NOTCH1 inhibitors do not present with 100% efficiency in the clinic,” Ntziachristos said.
Instead, targeting certain gene activating epigenetic modifications in these tumor-related neighborhoods restored normal DNA structure and gene regulation in models of disease, ultimately blocking tumor growth. This type of epigenetic inhibition could act as a “one-two punch,” by fighting cancer while leaving regular cells alone, according to Ntziachristos.
“We thus hope the inhibitors affect the growth of cancer cells, sparing physiological cells, but further research is warranted to test this hypothesis,” Ntziachristos said.
This study was supported by National Cancer Institute grants R00CA188293 and U54CA193419 (through the Chicago Region Physical Sciences–Oncology Center), the American Society of Hematology, the Zell Foundation and the H Foundation.