Discovery could help researchers design new chromatin-based therapeutics

Each cell in the human body holds a full two meters of DNA. In order for that DNA to fit into the cell nucleus — a cozy space just one hundredth of a millimeter wide — it needs to be packed extremely tight.
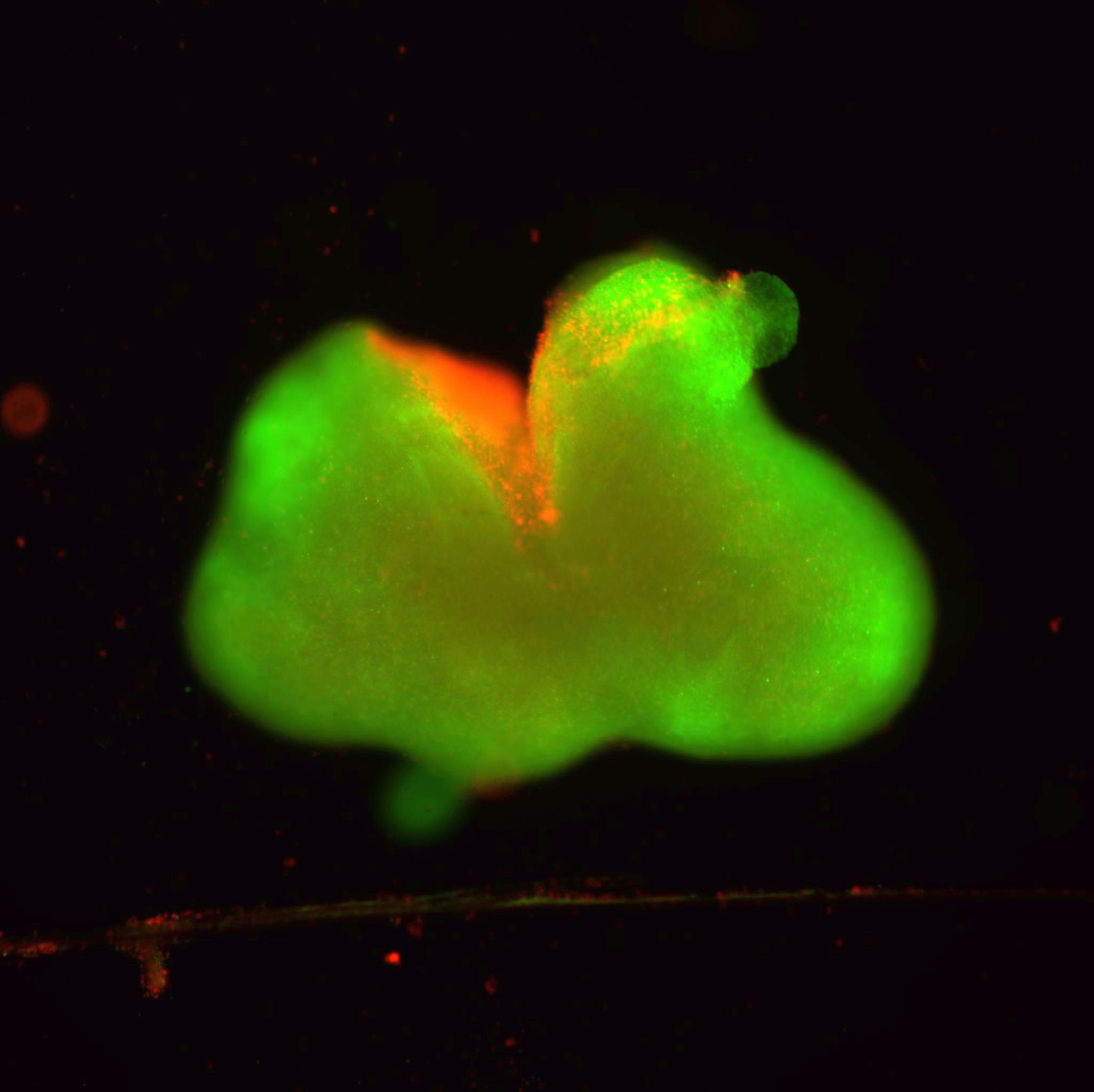
A new Northwestern University study published in Science Advances has discovered that the packing of the three-dimensional genome structure, called chromatin, controls how cells respond to stress. When the chromatin packing is heterogenous and disordered, a cell demonstrates more plasticity. When the packing is neat and orderly, a cell cannot respond as easily to outside stressors.
This discovery comes with both good and bad news.
The bad news: This means cancer cells with disordered packing are more likely to adapt to and evade treatments, such as chemotherapy. The good news: Now that researchers have this information, they can develop new cancer therapies that target chromatin packing. By inhibiting cancer cells’ ability to adapt, those cells become more vulnerable to traditional treatments.
“Cancer cells are masters of change,” said Northwestern’s Vadim Backman, PhD, who co-led the research. “They have to continuously adapt to evade the immune system, chemotherapies or immunotherapies. Abnormal chromatin packing drives cancer cells’ ability to do this.”
Backman is the Walter Dill Scott Professor of Biomedical Engineering in Northwestern’s McCormick School of Engineering and a professor of Biochemistry and Molecular Genetics, and of Medicine in the Division of Hematology and Oncology.
Backman, who is also the associate director for research technology and infrastructure at the at the Robert H. Lurie Comprehensive Cancer Center of Northwestern University, co-led the work with Igal Szleifer, PhD, the Christina Enroth-Cugell Professor of Biomedical Engineering, in McCormick, and of Medicine in the Division of Pulmonary and Critical Care, who is also a member of the Lurie Cancer Center.
Made of DNA, RNA and proteins, chromatin determines which genes get suppressed or expressed. In the case of cancer, chromatin can regulate expression of the genes that enable cells to become resistant to treatment.
“Genes are like hardware, and chromatin is software,” Backman said. “And chromatin packing is the operating system.”
“The genome has been sequenced, and techniques, such as CRISPR, now allow researchers to edit a cell’s ‘hardware,’” Szleifer added. “The role of chromatin structure on gene expression, however, has remained a scientific mystery.”
To help solve this mystery, Backman, Szleifer and their teams combined nanoscale imaging technologies with molecular dynamics modeling to analyze nanoscale alterations in chromatin packing structure.
Using data from The Cancer Genome Atlas, they analyzed cells from patients with colorectal, breast and lung cancers for markers of transcriptional plasticity. The researchers also used partial wave spectroscopic microscopy, which previously was developed in Backman’s laboratory, to examine chromatin in living cells in real time.
The researchers discovered an inverse relationship between patient survival and the plasticity of tumor cells. Backman says that chromatin engineering opens the door for a new class of cancer therapies, which could re-wire cells’ “operating systems” to make them less plastic.
“We found that transcriptional plasticity and chromatin packing alterations are an important marker, which is indicative of how a cancer patient may respond to anti-cancer therapies, such as chemotherapy,” said Ranya Virk, the paper’s first author. “This can be translated into a new method of predicting outcomes of cancer therapy.”
The study was supported by the National Science Foundation (award number EFMA-1830961) and the National Institutes of Health (award numbers R01CA228272, R01CA225002, U54CA193419 and T32GM008152). Northwestern PhD candidates Ranya Virk, Wenli Wu and Luay Almassalha were the paper’s co-first authors.