Northwestern Medicine scientists have discovered an epigenetic mechanism that preserves chromatin domains called centromeres during the process of DNA replication, according to a study published in Developmental Cell.
According to Daniel Foltz, PhD, associate professor of Biochemistry and Molecular Genetics, and senior author of the study, the findings shed light on a system that may lead to cancer when defective.
“Loss of centromeres contributes to chromosome instability by affecting chromosome number,” said Foltz, also a member of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University. “That can result in having too many or too few tumor suppressors, driving tumorigenesis, and lead to larger structural rearrangements of the chromosomes.”
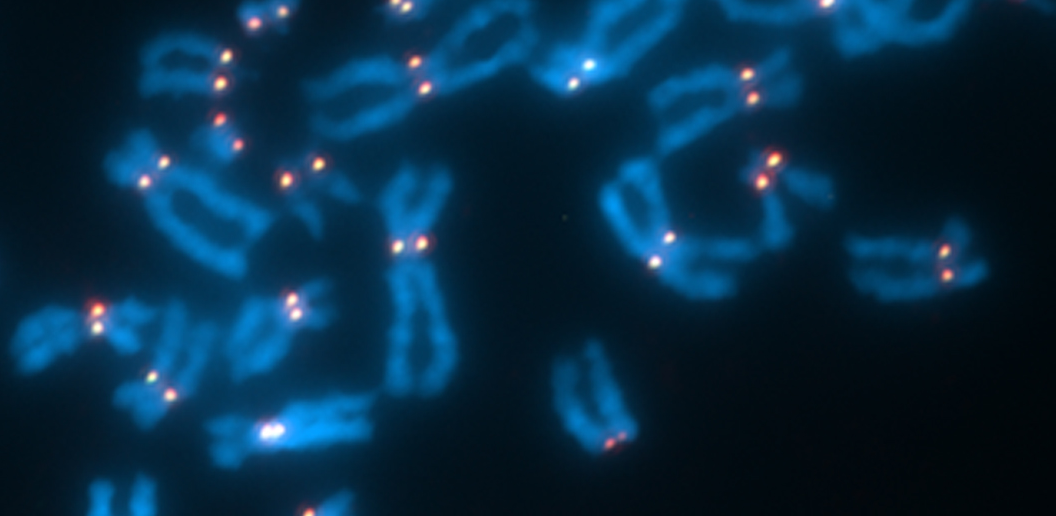
The centromere is a structure that provides a foundation for the segregation of duplicated chromosomes during cell division. The centromeres are located where the identical halves of the chromosome meet, creating an “X” shape. The centromere works to pull apart chromosomes during cell division, assembling a kinetochore that ensures equal segregation of sister chromosomes between daughter cells.
Each chromosome has a single site that is defined as a centromere and improper centromere placement can lead to misaligned chromosomes. Previous studies have shown this “centromere identity” is determined in part by a protein called CENP-A.
CENP-A is a histone protein that serves as an epigenetic marker. By building a nucleosome that resides at the centromere throughout the cell cycle, CENP-A ensures the centromere retains a stable position. However, it’s not that simple, according to Foltz.

“DNA replication requires the cell to melt the two pieces of DNA,” Foltz said. “The cell has to remove the histones from DNA before it can be replicated, creating an impediment to inheriting epigenetic information.”
Foltz and his colleagues hypothesized that there must be a mechanism for preserving this information and reassembling it after DNA replication. Using an unbiased screen for proteins bound to CENP-A during DNA replication, Foltz and colleagues discovered that a histone chaperone called HJURP facilitates CENP-A’s inheritance during DNA replication.
HJURP recognizes the CENP-A protein, plucking it out of the path of the DNA replication machinery and placing it back on the centromere once it’s reformed. This preserves CENP-A and by extension, centromeric identity, throughout DNA replication.
While major defects in this process simply lead to unviable cells, minor disturbance may trigger chromosomal instability — a common link between many cancers, according to Foltz.
“If you overexpress some known oncogenes, you perturb DNA replication,” Foltz said. “We’re interested in understanding how these perturbations may weaken the centromere and contribute to cancer.”
In addition, other epigenetic processes may be subject to these same inefficiencies, creating other opportunities for cancerous cells to proliferate.
“This has implications for epigenetic information that’s inherited through other histone variants and post-translational modifications outside of the centromere as well,” Foltz said. “Replication stress could lead to genomic instability that we know is a part of tumorigenesis.”
This study was supported by National Institutes of Health grants R01GM111907, R01GM082989 and F30CA186430; a Zell Scholar grant from the Robert H. Lurie Cancer Center and a pre-doctoral fellowship from the American Heart Association (15PRE25700271).