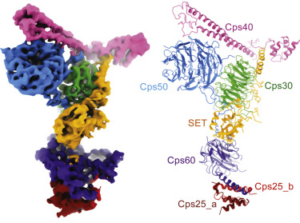
The three-dimensional atomic structure of the COMPASS protein complex has been detailed for the first time, according to a study published in the journal Cell.
These findings will provide a platform for designing future cancer therapies, as the epigenetic regulator COMPASS and members of its family are some of the most commonly mutated proteins in a variety of cancers, according to Yoh-hei Takahashi, PhD, research assistant professor of Biochemistry and Molecular Genetics and co-lead author of the study.
Ali Shilatifard, PhD, chair of Biochemistry and Molecular Genetics and the Robert Francis Furchgott Professor, was co-senior author of the study.
“We’ve got the blueprint to an engine: we know exactly where every screw and washer goes,” said Takahashi, who is a member of the Shilatifard laboratory. “Now we can open the engine up, learn about its activity and use this information to develop therapeutic targets.”

COMPASS, the first histone H3K4 methylase, was originally discovered in the Shilatifard laboratory over 20 years ago. Since then, Shilatifard, who is also the director of the Simpson Querrey Center for Epigenetics and a professor of Pediatrics, has been investigating COMPASS’ biological function, biochemical properties and gene expression and regulation. However, its complete physical structure remained unknown up until now.
In the current study, Takahashi biochemically reconstituted COMPASS, and in collaboration with Qianhui Qu, basic life research scientist at Stanford University, used cryo-electron microscopy to solve the atomic structure of COMPASS.
Qu is a member of the laboratory of Georgios Skiniotis, PhD, professor of Molecular & Cellular Physiology at Stanford University.
According to Shilatifard, cryo-electron microscopy is unique in that it allowed the scientists to examine the atomic structure of a large macromolecular complex while it was catalytically active; closer to its normal, functional state.

“That’s the key,” Shilatifard said. “You can get some polypeptide structures using X-ray but working with my colleague Dr. Skiniotis allowed us to obtain the structure of the whole complex with the catalytically active enzyme using cryo-electron microscopy.”
The investigators defined several important pieces of COMPASS, including a protein named Cps50 that was serving as a “molecular scaffold,” with structures extending to all other sub-units.
“This molecule is organizing the other components of the protein complex,” Takahashi said.
Another component, named Cps30, regulates catalytic function, which is important for cancer carcinogenesis. While the current study was performed in yeast, Cps30 has a human homologue that could be a prime target for therapy, according to Takahashi.
“By making a drug that targets Cps30’s domain interactions, that drug could alter the activity of COMPASS,” he said. “Changing the activity of COMPASS could kill cancer cells.”
In the future, Shilatifard and his collaborators will use the solved structure of COMPASS to investigate compounds that could regulate the stability of the complex, with the end goal of cancer therapy.
Joseph Brunzelle, PhD, research assistant professor at the Northwestern Synchrotron Research Center, was also a co-author on the study.
This work was supported by National Institutes of Health grant DK090165, Outstanding Investigator Award R35CA197569 from the National Cancer Institute and a Canadian Institutes of Health Research grant.