Northwestern Medicine scientists have revealed the role amino-terminal methylation plays in a specific protein in the centromere, a region of the chromosome important in cell division, and how the dysregulation of this protein can affect the development of cancer cells. Methylation of amino acid side chains is well-documented, but the role of amino-terminal methylation is much less well understood.
Published in Nature Communications, the study showed this posttranslational modification of the protein, centromere protein A (CENP-A), distinguishes it from a similar protein that is found in the rest of the chromosome. CENP-A is a type of histone, a protein with DNA wrapped around it, and specifies the location of the centromere in the nucleus.
Lead author Daniel Foltz, ’01 PhD, associate professor of Biochemistry and Molecular Genetics, and his team conducted functional analyses of the modifications on CENP-A, which they had previously identified.
“It’s interesting because this is a novel type of modifications on histones and because we can go in and really show what function is being mediated by the amino-terminal methylation, which has not been previously well-defined for this type of modification,” Foltz said.
The investigators discovered that CENP-A, when correctly methylated on its amino terminus, influences the recruitment of CCAN proteins, which are part of a large centromere complex of proteins. They showed that a subset of components of CCAN are dependent on this methylation. When the scientists blocked methylation from this process, they observed defects in chromosome segregation.
“We really defined a different arm of recruitment for the CCAN proteins than has been understood before,” Foltz said.
Overexpression of CENP-A in Cancer
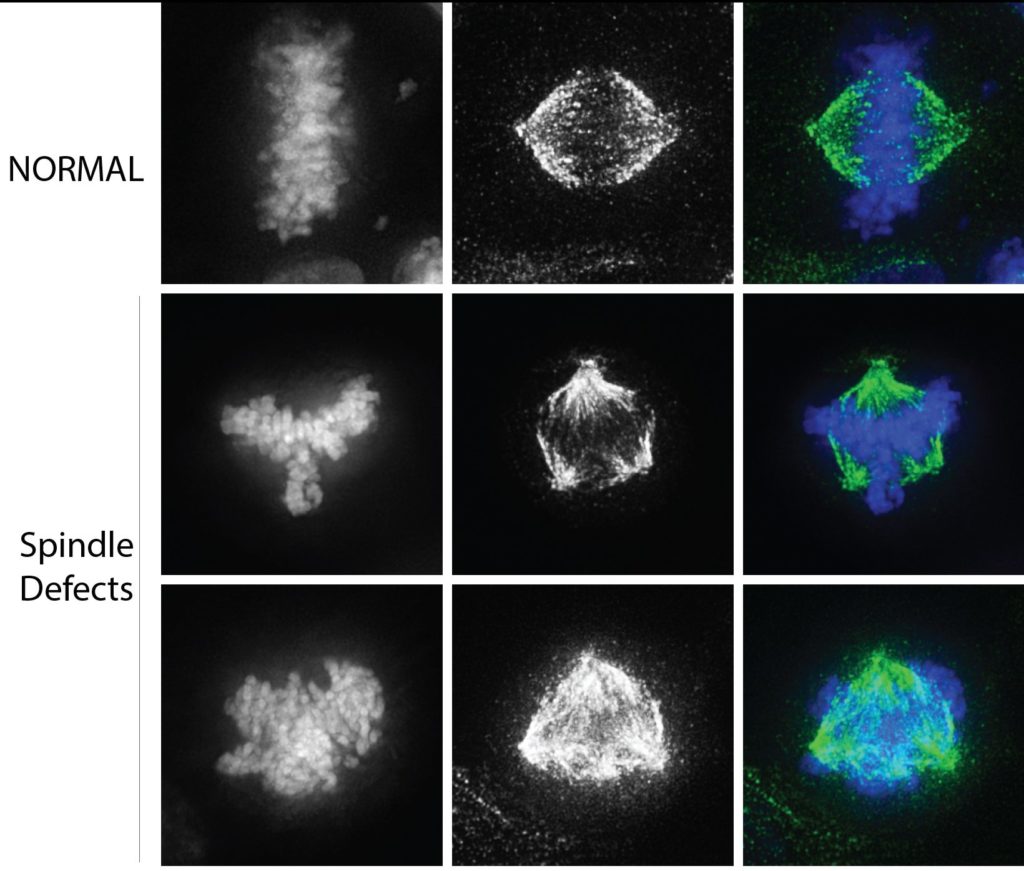
Next, the scientists studied hallmarks of cancer cells, including defects in chromosome segregation and spindle polarity. A bipolar spindle is essential to equally segregate chromosomes into two separate cells during cell division. In cancer cells, defects in the spindle lead to chromosomes that are pulled in multiple directions and can result in chromosome breakage and genomic instability. The scientists found that reducing the amount of methylation of CENP-A drives this process.
Foltz and the other investigators also found that in the absence of the tumor suppressor protein p53, loss of CENP-A methylation promotes more rapid tumor formation.
Next, Foltz and his team want to study how tumor cells are using this pathway.
“What we’ve done in this paper is engineer this defect into cells to see what the phenotype is,” Foltz said. “The next question is what happens to CENP-A methylation in cancer cells, and when CENP-A is overexpressed, to what degree is it driving the genomic instability in cancers?”
Foltz is also a member of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University.
The study was funded by Department of Defense Visionary Postdoctoral Fellowship W81XWH-13-1-0106, National Institutes of Health grant R01GM111907 and a research scholar award from the American Cancer Society.