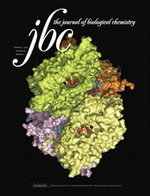
Scientists have revealed how a protein’s structural framework regulates the DNA building blocks required for a life-threatening hospital-borne pathogen to survive.
The bacterium, Enterococcus faecalis, undergoes frequent mutations, resulting in its dangerous antibiotic-resistant forms. To better understand its progression and adaptation, a team of investigators led by Wayne Anderson, PhD, professor in Molecular Pharmacology and Biological Chemistry, and Jinwoo Ahn, PhD, professor of Structural Biology at the University of Pittsburgh School of Medicine, studied EF1143, a protein responsible for maintaining the correct levels of DNA building blocks within cells.
“All organisms have to regulate the production of deoxyribonucleotide triphosphates (dNTPs) so that they are available in the right proportions to be used in the copying of DNA,” said Anderson, a member of the Center for Genetic Medicine, Center for Molecular Innovation and Drug Discovery and Robert H. Lurie Comprehensive Cancer Center. “If this balance is off, it can reduce the rate of DNA synthesis and increase the frequency of a mutation.”
The scientists conducted protein analyses of EF1143 and revealed its crystal structure, adding it to the National Institutes of Health (NIH) Protein Data Bank. The findings were featured on the cover of the Journal of Biological Chemistry.

“When we started working on this project, nothing was known about enzyme EF1143,” said Anderson, director of the Center for Structural Genomics of Infectious Diseases (CSIGD). Enzymes are proteins that speed up chemical reactions. “This paper, together with a preceding one, have described its structure, identified its enzymatic activity and discovered how the activity is regulated to maintain the appropriate concentrations of each of the four dNTPs needed for DNA synthesis.”
Determining protein structures lays the groundwork for drug discovery as they can help scientists identify potential new targets.
The study focused on E. faecalis, a bacterium found in the intestinal tract of humans, in an attempt to understand the molecular mechanisms of its development and adaptation to antibiotics. Several molecular components that make E. faecalis so dangerous had already been identified and the scientists concentrated on EF1143, which controls dNTP pools.
As part of a multi-institutional, international consortium that involves nine different laboratories across the United States, Canada and United Kingdom, investigators at the CSGID use x-ray crystallography and nuclear magnetic resonance to examine the atomic details of proteins from human pathogens.
The United Nations Educational, Scientific and Cultural Organization (UNESCO) has declared 2014 the International Year of Crystallography, in part to commemorate the first crystal structure determination in 1914. Crystallographic structure determination plays an important role in deciphering how biological macromolecules, like this protein, carry out their functions.
The work was supported in part by National Institutes of Health grants U54 GM094585 and P50GM082251.