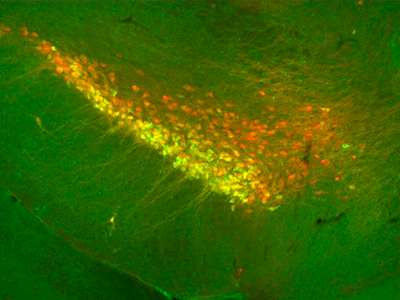
Though dopamine neurons influence many diverse behaviors and diseases, scientists have historically presumed that all of these important nerve cells are molecularly similar within two clusters of the brain. In a new study, Northwestern Medicine scientists prove that premise wrong by identifying several molecularly distinct subtypes of dopamine neurons within each cluster.
The finding, made possible with a new single-cell gene expression profiling method, could lead to better and more targeted therapies for diseases and disorders of the central nervous system.
The midbrain dopaminergic system – less than 1 percent of the brain’s neurons – controls many kinds of behaviors, including movement, appetite, reward, aversion and pleasure. And it’s involved in medical conditions such as Parkinson’s disease, schizophrenia, obsessive-compulsive disorder, ADHD, depression and drug addiction.
“How is it possible that such a small group of neurons accounts for all these different functions and diseases?” said senior author Rajeshwar Awatramani, PhD, associate professor in the Ken and Ruth Davee Department of Neurology and the Center for Genetic Medicine. “Within these clusters there must be different kinds of neurons, each potentially doing different things.”
The puzzle percolated in Awatramani’s mind for a decade, until “a fortuitous confluence of events” allowed him to investigate: First author Jean-Francois Poulin, PhD, a postdoctoral fellow with the same interest, joined his lab. At about the same time, new high-throughput technology became available.
“A company called Fluidigm developed a microfluidic array that allowed us to take single dopaminergic neurons and look at their gene expression profiles in a very cost-effective and fast way,” said Awatramani.
Most midbrain dopamine neurons cluster in the substantia nigra or the ventral tegmental area of the brain. Previous studies presumed that neurons in one structure are different from those in the other, but that all the neurons within each are the same. Therefore, they conducted gene expression analysis on the whole samples rather than individual cells.

By profiling single cells, 96 genes at a time, Awatramani and Poulin discovered that the two main clusters contain multiple intermingled subtypes of neurons. Their results were published in Cell Reports.
“Some of these cell types have very unique markers, molecular signatures that give us genetic access to those populations,” said Awatramani. “With that information, we can develop tools to shut off a type of neuron in mouse models and find out its function.”
Ultimately, this study’s findings could make it possible to target specific neuron subtypes – instead of all of them – in therapies for Parkinson’s and other diseases.
“The adverse effects of dopaminergic therapies could possibly be minimized by knowledge of what’s expressed in each of these subsets,” said Awatramani, who is a member of the Robert H. Lurie Comprehensive Cancer Center.
In addition to dopamine neurons, he says that scientists can use the same microfluidic array method to reclassify other neurotransmitter populations in the brain.
“Classifying neurons in the brain is one of the main goals of the Obama administration’s BRAIN Initiative,” he said. “This single cell platform is a very nice way to achieve that goal.”
This work was supported by the Paul Ruby Foundation for Parkinson’s Research through the through Northwestern Memorial Foundation; Fonds de recherche du Québec – Santé; Michael J. Fox Foundation; Canadian Institutes of Health Research; National Institutes of Health (NIH) National Institute of Neurological Disorders and Stroke grant 1R21NS072703-01; and NIH Clinical and Translational Science Awards UL1 TR000150 and UL1 RR025741 to Feinberg.