A multidisciplinary team of investigators has developed a first-of-its-kind interactive 3D spatial approach that reveals new therapeutic targets and provides users with a comprehensive three-dimensional view of glioblastoma tumors, detailed in a recent study published in Cell.
The approach provides a whole-tumor analysis on glioblastoma, the most lethal primary brain tumor in adults, and highlights tumor-wide features that may inform the development of new precision medicine strategies.
“This is the first such type of study in brain tumors or in any solid tumor where you can study genetic mutations across the whole tumor rather than focusing on just one piece of the tumor,” said Feng Yue, PhD, the Duane and Susan Burnham Professor of Molecular Medicine and co-senior author of the study.
Glioblastoma is an aggressive and treatment-resistant brain tumor with poor prognosis. Patients’ average length of survival after diagnosis is an estimated 15 months and the average rate of five-year survival is less than seven percent.
Poor treatment outcomes have been partially attributed to the intratumor heterogeneity and evolution, which helps the tumor resist treatments.
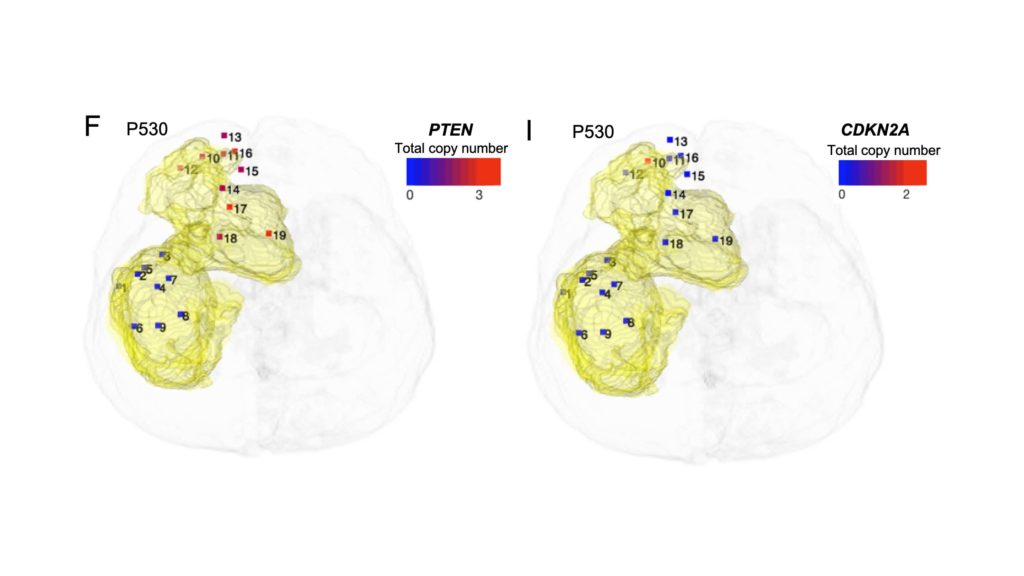
However, defining these diverse characteristics throughout the entire glioblastoma tumor, as well as all other types of solid tumors, has remained a challenge as research has historically relied on single-tissue biopsies that can characterize only one area of the tumor at a time.
“Traditionally, to find genetic mutations in tumors, people just take one piece from the tumor and perform target or whole genome sequencing. But because there could be different mutations in different parts of the tumor, such approaches might miss the potential key driver mutations, so that’s why we really want to study the mutations at the whole-tumor level,” said Yue, who is a professor of Biochemistry and Molecular Genetics, of Pathology and director of the Center for Advanced Molecular Analysis.
In response, a multidisciplinary team of investigators including Yue developed a novel three-dimensional neuro-navigation platform that can help identify intratumoral features and spatial patterns in multiple regions of a patient’s glioblastoma tumor at once.

The investigators then used their platform with tumor-tissue samples from 10 patients with newly diagnosed and untreated glioblastoma and produced more than 100 spatially mapped samples highlighting tumor diversity.
Coordinates from these samples were then integrated with MRI scans of the patient’s brains to create an interactive three-dimensional, 360-degree map of the patient’s entire tumor.
Then, Yue and his collaborators interrogated these samples with a variety of genomics, epigenomics and single cell approaches to reveal the tumors microenvironmental heterogeneity and spatial patterns, enabling the investigators to identify the earliest origins and clonal evolution of the tumors.
Furthermore, in their 3D system, users can select which features in the tumor to highlight, including genomic, epigenomic and microenvironmental mutations, which can help scientists better understand the evolutionary molecular pathways that the tumor may have used to become treatment resistant, according to Yue.
“The idea is that if we find one mutation presenting in all different spots of the same tumor, that mutation is more likely to be the driver mutation or the cancer initiation event. But if we only study one spot of the tumor, we probably won’t have such a clear view of the clonal initiation and evolution process,” Yue said.
The study is the first to display glioblastoma evolution and heterogeneity from a three-dimensional whole-tumor perspective, and may help identify novel therapeutic targets in which current precision therapies have otherwise failed to effectively treat due to tumor heterogeneity, according to Yue.
“Through these types of studies, we’re able to identify many novel fusions that have been missed by other platforms. Some of them can even be targeted by FDA-approved drugs,” Yue said.
Co-authors of the study include Qixuan Wang, a student in the Driskill Graduate Program in Life Sciences (DGP); Qiushi Jin; Lena Stasiak; Ye Hou; Juan Wang, PhD, a postdoctoral fellow in the Yue laboratory; and Mark Youngblood, MD, PhD, a resident in the Department of Neurological Surgery.
Yue is also a member of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University and director of the Center for Cancer Genomics.
This work was a collaboration between Yue’s group and a team of investigators from the University of California San Fransisco, led by Joseph Costello, PhD. This work was supported in part by National Institutes of Health grants R35GM124820, 1RO1HG009906 and RO1HG11207.