A recent Northwestern Medicine study published in the Journal of Cell Biology has identified new mechanisms that cause genomic or chromosomal instability during cell division, findings that may improve the development of biomarkers and targeted therapies for cancer.
For cells to successfully proliferate, chromosome segregation must occur following DNA replication. However, this process can also result in genomic instability, which occurs when chromosomes are improperly distributed during cell division, or mitosis, resulting in compromised cellular function.
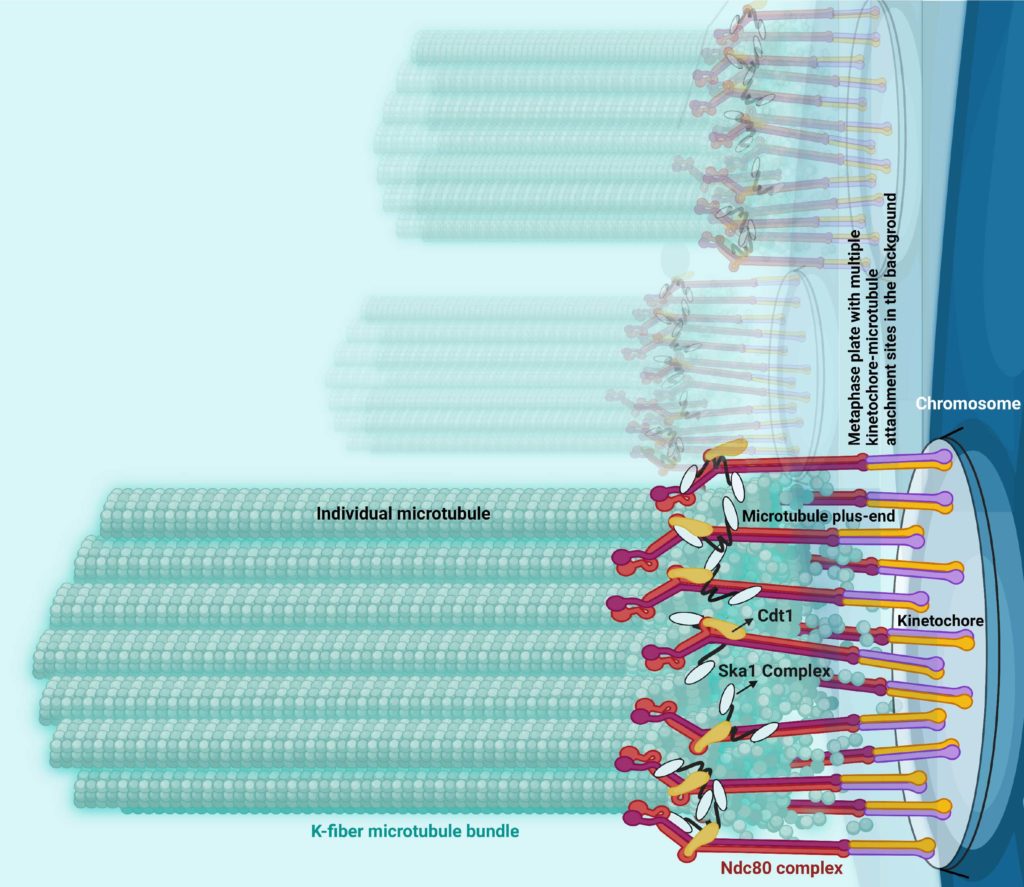
A football-shaped structure called the mitotic spindle is assembled at the beginning of mitosis, serving as a framework for the events of chromosome segregation. When cells proliferate properly, chromosomes get condensed and they attach to the ends of mitotic spindle microtubules, a key component of the cell’s cytoskeleton that supports cell division as well as cell migration, intracellular transport and cell structure.
“There is instability at the ends of spindle microtubules in that tubulin protein monomers are being added and removed constantly, so it’s not a static structure, and then you need the chromosomes to attach to that dynamic structure. That’s a pretty challenging task and most importantly, the mechanisms of how that happens in human cells is not known,” said Dileep Varma, PhD, assistant professor of Cell and Developmental Biology and senior author of the study.
Previous work has established the protein Ndc80 as essential for chromosomes to attach to microtubules and two additional proteins — Cdt1 and Ska1 — separately help Ndc80 bind, or “couple,” to the ends of the microtubules. This “coupling,” according to Varma, is crucial because otherwise chromosomes get persistently detached from microtubule ends, which can lead to harmful cellular mutations or gene dosage problems brought about by chromosome gain or loss.
“Forming an attachment is not enough; it needs to be stably coupled to the ends of these microtubules, and if the chromosomes don’t attach to this structure, they don’t segregate properly into the two daughter cells. That’s when you lose the chromosomes, and then that’s when you have chromosomal instability which is a key driver for cancer,” said Varma, who is also a member of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University.
In the current study, Varma’s team developed a novel approach which involves attaching a degron, an amino acid sequence “tag,” to the end of the desired proteins and then, using CRISPR genome editing, replacing the desired gene on the chromosome with copies of the same gene with the degron attached to it.
This approach allowed the investigators to precisely inhibit the Cdt1 protein function during mitosis without interfering with its DNA replication function, according to Varma.
In addition to functioning during chromosome-microtubule coupling, Cdt1, has also been shown to be important for DNA replication.
Using this and other novel approaches, the investigators discovered that during chromosome segregation and mitosis, the protein Ska1 guides Cdt1 to the ends of the microtubules to bind to this structure, and then both these proteins join with Ndc80 at kinetochores — protein structures that connect chromosomes to microtubules — to form a larger complex that constitutes the central chromosome-microtubule coupling unit.

“The formation of this tripartite complex consisting of Ndc80, Cdt1 and Ska1 enables kinetochores to robustly couple the microtubule ends, and that is really what causes this persistent coupling of chromosomes for longer durations and for longer distances without detachment within a mitotic cell,” Varma said.
According to Varma, his team’s findings provide further insight into the complexity of cell division and could help improve the development of biomarkers and targeted therapies for cancer, such as small molecule inhibitors.
“When you target a protein like Cdt1 for cancer therapy, there are certain unique aspects of the protein that helps it be a better candidate than not, and that is essentially the availability or presence of smaller structural domains that you can develop drugs against to inhibit,” Varma said.
Amit Rahi, PhD, a post-doctoral fellow in the Varma laboratory, was co-first author of the study.
This work was supported by National Institute of General Medical Sciences grant R01GM135391.