A Northwestern Medicine study has identified a protein that drives breast cancer stemness and metastasis. The findings, enabled by machine learning, were published in the journal eLife.
Tumor-initiating cells with stem-progenitor cell properties (stemness) are thought to be essential for cancer development and metastatic regeneration in many cancers. However, elucidation of the underlying molecular network has been challenging, according to the study’s senior author Huiping Liu, MD, PhD, professor of Pharmacology and of Medicine in the Division of Hematology and Oncology.
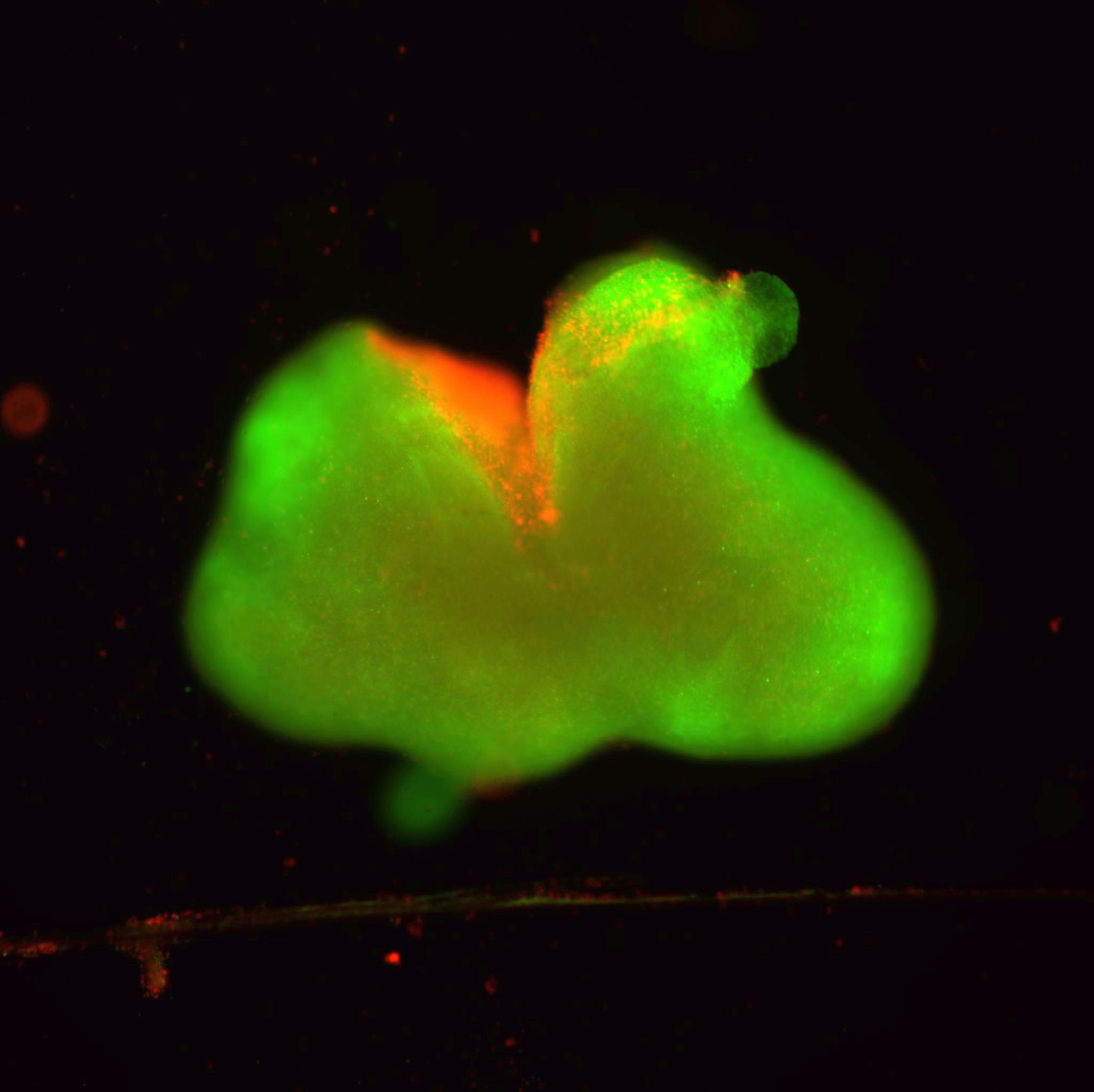
Combining machine learning and experimental investigation, Liu and her team demonstrated that the protein membrane CD81 interacts with another previously identified tumor-initiating cell marker, called CD44, in promoting tumor cell cluster formation and lung metastasis of triple negative breast cancer (TNBC).
“The findings have broadened the knowledge of the molecular regulatory network of breast cancer cell stemness and may be significant for therapeutic targeting,” Liu said.
TNBC comprises 10 to 15 percent of newly diagnosed breast cancer cases and is highly metastatic with low long-term survival. TNBC metastasizes to the visceral organs, such as the lungs, liver and brain. While the cellular mechanisms contributing to the spreading of the tumor cell remain largely unknown, protein networks offer a clue, according to Liu.
“We know that protein functions and their networks are part of the backbones directly mediating cellular phenotypes and performance,” said Liu, who is also a member of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University.
In previous research, Liu’s laboratory found that the breast tumor-initiating cell marker CD44 mediates formation of circulating tumor cell (CTC) clusters (two or more cells) and its enriched expression in such clusters predicts an unfavorable overall survival of patients with breast cancer, especially TNBC. CTC clusters possess up to 100-fold greater efficiency than single CTCs in seeding metastasis.
In this study, to characterize the protein network of CD44, the scientists found that CD44 binds to CD81 for coordinated membrane localization, and both are required for optimal self-renewal, CTC cluster formation, and metastasis, emphasizing the indispensable functions of CD44 and CD81 in cell adhesion and intercellular interactions in metastasis, Liu said.
To demonstrate the partnership between the two membrane proteins, the scientists used a sophisticated, computational algorithms-based protein structural modeling (machine learning) to predict the interface regions and amino acid residues involved in the protein interactions. Experimental approaches were then used to validate CD44 and CD81 interface interactions.
“Machine learning and deep learning have transformed protein structure modeling, greatly facilitating the molecular understanding and therapeutic development for TNBC and other metastatic disease,” Liu said.
Liu adds that machine learning and deep learning-facilitated research will not only exponentially scale up the simulated therapeutic screening for best efficacy and lowest toxicity, but also make personalized medicine more achievable and affordable.
“Our next step is to combine the CTC biomarker analysis with personalized targeting therapy development,” she said.
Tujin Shi, PhD, a senior scientist at Pacific Northwest National Laboratory and Yang Shen, PhD, at Texas A&M University were co-senior authors of this study, contributing to proteomic analyses and machine learning, respectively for CD81-CD44 network and molecular signaling studies. Massimo Cristofanilli, former professor of Medicine in the Division of Hematology and Oncology (now at Cornell University), provided patient blood samples to the CTC analyses in the study. Erika Ramos, a student in the Driskill Graduate Program in Life Sciences (DGP), was the first author of the study and research associate Nurmaa Dashzeveg was the co-last author; both are in the Liu laboratory.
This research was supported by Department of Defense grants W81XWH-16-1-0021 and W81XWH-20-1-0679; NIH/NCI grants R01CA245699, R35GM124952 and R01GM139858; National Science Foundation CCF-1943008; the Lynn Sage Cancer Research Foundation; Susan G. Komen Foundation CCR18548501; American Cancer Society ACS127951-RSG-15-025-01-CSM; Northwestern University, NIH Fellowships T32 CA009560 and T32 CA080621-15; and the Julius Kahn Fellowship and T32GM008061.