Investigators have discovered the mechanisms underlying a T-cell receptor’s immunosuppressive function, according to a study published Nature Immunology, findings that may provide insight into the development of novel precision therapeutics for chronic diseases, including cancer.
Hui Zhang, PhD, professor of Preventive Medicine in the Division of Biostatistics, a member of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University, and director of Lurie Cancer’s Brain Tumor SPORE Biostatistics and Bioinformatics Core and of the Biostatistics and Data Management Core in the Mesulam Center for Cognitive Neurology and Alzheimer’s Disease, was a co-author of the study.
LAG3 is an immune checkpoint receptor expressed on the surface of exhausted T-cells, which progressively lose their immune functions while fighting chronic infection or cancer. As an immune checkpoint, LAG3 inhibits the activation of the host cell and promotes an immunosuppressive response.
While immunotherapy treatments targeting LAG3 continue to be evaluated in clinical trials, the mechanisms by which LAG3 inhibits T-cell function and proper immune response have remained understudied.
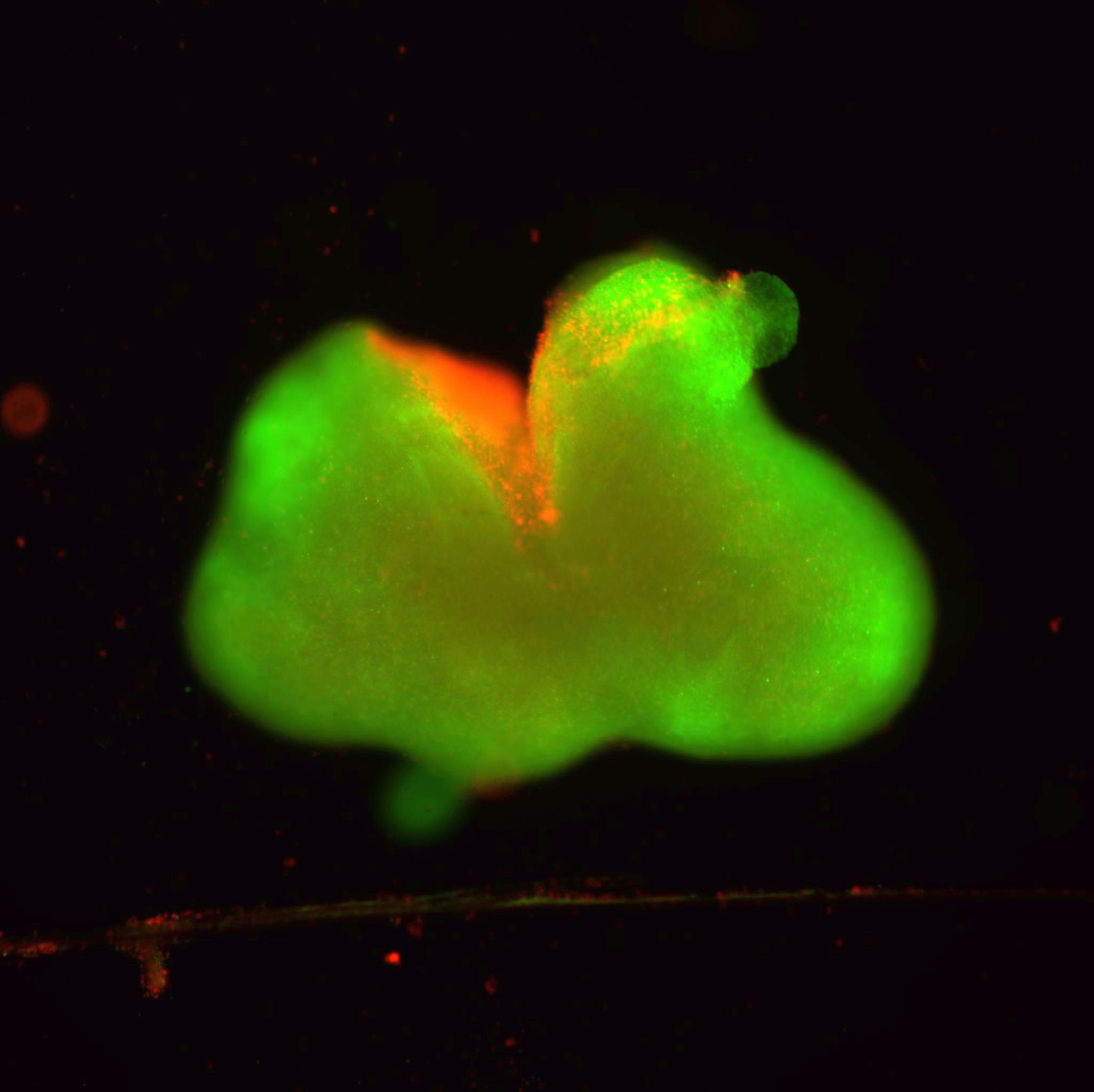
In the current study, investigators analyzed CD4+ and CD8+ T-cells in mice, and discovered that LAG3 relocates to the immunological synapse — the space between an antigen-presenting cell and the immune cell — and binds with the T-cell receptor (TCR) CD3 protein complex.
The investigators noted this relocation occurred instead of LAG3 binding to major histocompatibility complex (MHC) class II molecules, which are found on antigen-presenting cells and are essential for initiating a proper immune response.
Using two super-resolution microscopy techniques — stimulated emission depletion (STED) microscopy and stochastic optical reconstruction microscopy (STORM) — the investigators found that LAG3 colocalized with the TCR–CD3 complex in CD4+ and CD8+ T-cells, resulting in the loss of proper TCR signaling and decreasing T-cell activation.
To identify and quantify the coordinate-based single-molecule data from STED and STORM, investigators utilized an innovative statistical method proposed by Zhang’s laboratory, the normalized spatial intensity correlation (NSInC) statistical algorithm, which allows for unbiased analysis of coordinate-based single-molecule data to determine which identified proteins are colocalized in three-dimensional space.
“These observations indicated that LAG3 functioned as a signal disruptor in a major histocompatibility complex class II-independent manner and provide insight into the mechanism of action of LAG3-targeting immunotherapies,” the authors wrote.
Because current LAG3-targeting cancer therapeutics block the LAG3–MHC class II interaction, the findings may aide in the development of therapeutics targeting LAG3 and the LAG3–TCR association, as well as for the treatment of autoimmune and inflammatory diseases, according to the authors.
This work was supported by the National Institutes of Health grants P01 AI108545, R01 AI129893 and R01 AI144422; National Cancer Institute Comprehensive Cancer Center Support CORE grant CA047904; and the American Lebanese Syrian Associated Charities.