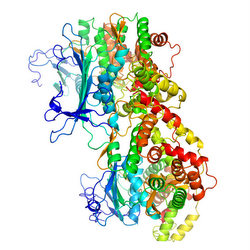
On May 30, the Center for Structural Genomics of Infectious Diseases deposited seven structures in the Protein Data Bank, pushing it past the 500 structure milestone. Among them was an aminoglycoside phosphotransferase (APH) in complex with the antibiotic kanamycin and a protein kinase inhibitor. This enzyme provides bacteria resistance to kanamycin, an APH antibiotic used to treat a wide variety of infections. One possibility for overcoming the resistance is identify a drug that can inhibit the enzyme.
Researchers at the Center for Structural Genomics of Infectious Diseases (CSGID) are well attuned with making blueprints – though they’re often filled with hues of red, green, and orange, too.
As part of the multi-institutional, international consortium that involves nine different laboratories across the United States, Canada, and United Kingdom, investigators at Northwestern University Feinberg School of Medicine are using X-ray crystallography and nuclear magnetic resonance to examine the atomic details of proteins from human pathogens.
Although the colorful images of these three-dimensional atomic structures are attractive in their own right, the goal of the project is to have scientists make use of them as they combat deadly infectious diseases.
“We are laying the groundwork for drug discovery,” said Wayne Anderson, PhD, professor in molecular pharmacology and biological chemistry, and director of the CSGID. “Determining protein structures can help researchers find potential targets for new drugs, essential enzymes, and possible vaccine candidates.”
Since the first structure was placed in the National Institutes of Health-supported Protein Data Bank (PDB) in April 2008, the CSGID and its counterpart, the Seattle Structural Genomics Center for Infectious Disease (SSGCID), have each added more than 500 others.
The 1,000 protein milestone was one that seemed far off after the first 12 months of the five-year, $31 million contract from the National Institute of Allergy and Infectious Diseases at NIH. With very few structures completed, it was hard to imagine a time when seven structures might be uploaded to the PDB on a single day, as was the case May 30.
“It used to take four years to determine one structure,” Anderson said. “Now we can finish about three per week, and we are continually working on more than 6,000 approved targets.”
Operating at a pace of about 150 structures per year, the CSGID has helped examine the atomic details of proteins from more than 40 human pathogens, including those responsible for the plague, anthrax, salmonellosis, cholera, tuberculosis, dengue fever, and influenza.
Selected for their biomedical relevance, as well as potential therapeutic and diagnostic benefits, one-third of the proteins are direct requests from the infectious disease research community. Scientists interested in nominating protein targets for structural determination can complete a request form on the CSGID or SSGCID website, according to their organism of interest. Both centers also freely distribute the protein expression clones via the NIH-funded Biodefense and Emerging Infections Research Resources Repository.
Each institution in the consortia contributes to different aspects of the high-throughput pipeline, which include selecting target proteins using bioinformatics, cloning their genes into bacteria for protein production, purification, and crystallization, with X-ray diffraction data collected at the Advanced Photon Source synchrotron at Argonne National Laboratory. After the protein structures are solved, their coordinates are made available freely to the research community by deposition into the PDB.
“In addition to the aim of providing a starting point for structure-based drug discovery, we can also use this research as a way to learn more about these pathogens and how they cause diseases, how they get around the immune system, how we defend ourselves against them and how they interact with us or their host,” Anderson said.
One of the major challenges in medicine today is fighting bacteria that have become drug-resistant. Methicillin-resistant Staphylococcus aureus (MRSA) is incredibly difficult to treat because it has developed a resistance to antibiotics, including penicillin.
“By determining the structure of proteins targeted by these drugs, we can now look at how the atoms are arranged in space and how they interact with one another,” Anderson said. “Then researchers can determine how the bacterium developed resistance and figure out what to change in the drug so that the bacteria will not recognize it.”
CSGID has been funded in whole or in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institute of Health, Department of Health and Human Services, under contract numbers HHSN272200700058C.
Those interested in collaborating with CSGID and writing publications about the many 3D structures deposited in the PDB can contact Anderson or project manager Elisabetta Sabini.